SF3B1 mutations constitute a novel therapeutic target in breast cancer
- PMID: 25424858
- PMCID: PMC4643177
- DOI: 10.1002/path.4483
SF3B1 mutations constitute a novel therapeutic target in breast cancer
Abstract
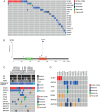
Mutations in genes encoding proteins involved in RNA splicing have been found to occur at relatively high frequencies in several tumour types including myelodysplastic syndromes, chronic lymphocytic leukaemia, uveal melanoma, and pancreatic cancer, and at lower frequencies in breast cancer. To investigate whether dysfunction in RNA splicing is implicated in the pathogenesis of breast cancer, we performed a re-analysis of published exome and whole genome sequencing data. This analysis revealed that mutations in spliceosomal component genes occurred in 5.6% of unselected breast cancers, including hotspot mutations in the SF3B1 gene, which were found in 1.8% of unselected breast cancers. SF3B1 mutations were significantly associated with ER-positive disease, AKT1 mutations, and distinct copy number alterations. Additional profiling of hotspot mutations in a panel of special histological subtypes of breast cancer showed that 16% and 6% of papillary and mucinous carcinomas of the breast harboured the SF3B1 K700E mutation. RNA sequencing identified differentially spliced events expressed in tumours with SF3B1 mutations including the protein coding genes TMEM14C, RPL31, DYNL11, UQCC, and ABCC5, and the long non-coding RNA CRNDE. Moreover, SF3B1 mutant cell lines were found to be sensitive to the SF3b complex inhibitor spliceostatin A and treatment resulted in perturbation of the splicing signature. Albeit rare, SF3B1 mutations result in alternative splicing events, and may constitute drivers and a novel therapeutic target in a subset of breast cancers.
Keywords: SF3B1; alternative splicing; breast cancer; drivers; next-generation sequencing; spliceostatin A.
© 2014 The Authors. The Journal of Pathology published by John Wiley & Sons Ltd on behalf of Pathological Society of Great Britain and Ireland.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

