New functions and signaling mechanisms for the class of adhesion G protein-coupled receptors
- PMID: 25424900
- PMCID: PMC4278406
- DOI: 10.1111/nyas.12580
New functions and signaling mechanisms for the class of adhesion G protein-coupled receptors
Abstract
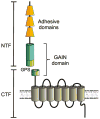
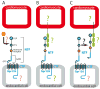
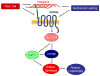
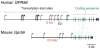
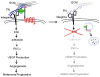
The class of adhesion G protein-coupled receptors (aGPCRs), with 33 human homologs, is the second largest family of GPCRs. In addition to a seven-transmembrane α-helix-a structural feature of all GPCRs-the class of aGPCRs is characterized by the presence of a large N-terminal extracellular region. In addition, all aGPCRs but one (GPR123) contain a GPCR autoproteolysis-inducing (GAIN) domain that mediates autoproteolytic cleavage at the GPCR autoproteolysis site motif to generate N- and a C-terminal fragments (NTF and CTF, respectively) during protein maturation. Subsequently, the NTF and CTF are associated noncovalently as a heterodimer at the plasma membrane. While the biological function of the GAIN domain-mediated autocleavage is not fully understood, mounting evidence suggests that the NTF and CTF possess distinct biological activities in addition to their function as a receptor unit. We discuss recent advances in understanding the biological functions, signaling mechanisms, and disease associations of the aGPCRs.
Keywords: adhesion G protein-coupled receptor; cancer; development; myelination; signal transduction; structural biology; synaptogenesis.
© 2014 New York Academy of Sciences.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Arac D, et al. Dissecting signaling and functions of adhesion G protein-coupled receptors. Ann N Y Acad Sci. 2012;1276:1–25. - PubMed
-
- Fredriksson R, et al. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. - PubMed
-
- Krishnan A, et al. Remarkable similarities between the hemichordate (Saccoglossus kowalevskii) and vertebrate GPCR repertoire. Gene. 2013;526:122–133. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

