The domestication of the probiotic bacterium Lactobacillus acidophilus
- PMID: 25425319
- PMCID: PMC4244635
- DOI: 10.1038/srep07202
The domestication of the probiotic bacterium Lactobacillus acidophilus
Abstract
Lactobacillus acidophilus is a Gram-positive lactic acid bacterium that has had widespread historical use in the dairy industry and more recently as a probiotic. Although L. acidophilus has been designated as safe for human consumption, increasing commercial regulation and clinical demands for probiotic validation has resulted in a need to understand its genetic diversity. By drawing on large, well-characterised collections of lactic acid bacteria, we examined L. acidophilus isolates spanning 92 years and including multiple strains in current commercial use. Analysis of the whole genome sequence data set (34 isolate genomes) demonstrated L. acidophilus was a low diversity, monophyletic species with commercial isolates essentially identical at the sequence level. Our results indicate that commercial use has domesticated L. acidophilus with genetically stable, invariant strains being consumed globally by the human population.
Conflict of interest statement
M.B. 's industrial CASE PhD studentship was partly sponsored by the Cultech Ltd, who manufacture nutritional supplements including probiotic products; the remaining authors have no conflicts of interests to declare.
Figures

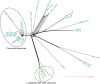
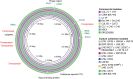
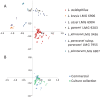
References
-
- Kandler O. Carbohydrate metabolism in lactic acid bacteria. Antonie van Leeuwenhoek 49, 209–224, 10.1007/BF00399499 (1983). - PubMed
-
- Felis G. E. & Dellaglio F. Taxonomy of Lactobacilli and Bifidobacteria. Curr Issues Intestinal Microbiol 8, 44–61 (2007). - PubMed
-
- Euzeby J. P. List of Bacterial Names with Standing in Nomenclature: a folder available on the Internet. Int J Syst Bacteriol 47, 590–592 (1997). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

