Cytosolic RNA:DNA hybrids activate the cGAS-STING axis
- PMID: 25425575
- PMCID: PMC4282641
- DOI: 10.15252/embj.201488726
Cytosolic RNA:DNA hybrids activate the cGAS-STING axis
Abstract
Intracellular recognition of non-self and also self-nucleic acids can result in the initiation of potent pro-inflammatory and antiviral cytokine responses. Most recently, cGAS was shown to be critical for the recognition of cytoplasmic dsDNA. Binding of dsDNA to cGAS results in the synthesis of cGAMP(2'-5'), which then binds to the endoplasmic reticulum resident protein STING. This initiates a signaling cascade that triggers the induction of an antiviral immune response. While most studies on intracellular nucleic acids have focused on dsRNA or dsDNA, it has remained unexplored whether cytosolic RNA:DNA hybrids are also sensed by the innate immune system. Studying synthetic RNA:DNA hybrids, we indeed observed a strong type I interferon response upon cytosolic delivery of this class of molecule. Studies in THP-1 knockout cells revealed that the recognition of RNA:DNA hybrids is completely attributable to the cGAS-STING pathway. Moreover, in vitro studies showed that recombinant cGAS produced cGAMP upon RNA:DNA hybrid recognition. Altogether, our results introduce RNA:DNA hybrids as a novel class of intracellular PAMP molecules and describe an alternative cGAS ligand next to dsDNA.
Keywords: RNA:DNA hybrids; STING; cGAS; innate immunity; pattern recognition receptor.
© 2014 The Authors.
Figures
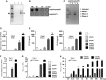
Synthetic poly(rA) (lane 1) and poly(dT) (lane 2) oligos were annealed, and the samples were run on a 1% agarose gel. Arrows indicate the annealed poly(rA):poly(dT) (lane 3) RNA:DNA hybrid.
Different amounts of synthetic annealed poly(rA):poly(dT) hybrid was spotted on a nitrocellulose membrane and probed with anti-hybrid S9.6 antibody.
Poly(rA):poly(dT) was either undigested (1) or digested with DNase I (2), RNase A (3) or RNase H (4), and the digested samples were analyzed on a 1% agarose gel.
Human peripheral blood mononuclear cells (PBMCs) were transfected with or without poly(rA), poly(dT) or poly(rA):poly(dT) hybrid or poly(dA):poly(dT) dsDNA for 6 h. RT–PCR was performed to check for the expression of IFNB. Figure shows values from two biological replicates. Data plot shows mean with SEM.
Human PBMCs from two different donors were transfected with or without poly(rA), poly(dT) or poly(rA):poly(dT) for 16 h, and the amount of secreted IP-10 was measured by ELISA. Each experiment was performed in duplicate. Data plot shows mean with SEM.
Differentiated human monocyte THP-1 cells were transfected with or without poly(rA):poly(dT) hybrid, poly(dA):poly(dT) dsDNA and RT–PCR was performed to check for the expression of IFNB. Each experiment was performed in duplicate. Data are representative of one of two independent experiments. Data plot shows mean with SEM.
Differentiated human monocyte THP-1 cells with Gaussia luciferase knockin under IFIT1 promoter were transfected with or without poly(rA):poly(dT) hybrid, poly(dA):poly(dT) dsDNA and luciferase assay was performed. Data shown are from two biological replicates. Data plot shows mean with SEM.
Differentiated human monocyte THP-1 cells with Gaussia luciferase knockin under IFIT1 promoter were transfected with different amounts of poly(rA):poly(dT) hybrid or poly(dA):poly(dT) dsDNA and luciferase assay was performed. Data shown are from two biological replicates. Data plot shows mean with SEM.
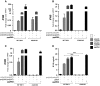
A–C WT THP-1, MAVS KO THP-1, STING KO THP-1 and cGAS KO THP-1 cells were transfected with or without poly(rA):poly(dT) (hybrid), poly(dA):poly(dT) dsDNA or pppRNA for 6 h. RT–PCR was performed to check for the expression of IFNB. Data shown are from experimental duplicates representative of two independent experiments. Data plot shows mean with SEM.
D WT THP-1 cells or cGAS KO THP-1 cells were transfected with or without poly(rA):poly(dT) (hybrid), dsDNA or pppRNA for 16 h, and amount of IP-10 secreted was detected by ELISA. Data shown are from experimental triplicate, representative of three independent experiments. Data plot shows mean with SEM.
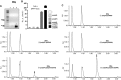
60-bp synthetic poly(rA) (lane 1) and 60-bp poly(dT) (lane 2) oligos were annealed, and the samples were run on a 1% agarose gel. Asterisk indicates annealed poly(rA):poly(dT) RNA:DNA hybrid (lane 3).
Differentiated human monocyte THP-1 cells with Gaussia luciferase knockin under IFIT1 promoter were transfected with or without 60-bp poly(rA), 60-bp poly(dT) or 60-bp poly(rA):poly(dT) (hybrid) and 60-bp poly(dA):poly(dT) (dsDNA) and luciferase assay was performed. Data shown are from two biological replicates. Data plot shows mean with SEM.
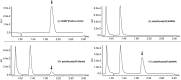
RP-HPLC chromatograms showing the presence of cGAMP(2′–5′) after incubation of recombinant cGAS protein for 180 min in the presence of ATP and GTP with: (i) 60-bp poly(rA) (ssRNA), (ii) 60-bp poly(dT) (ssDNA), (iii) 60-bp poly(dA) (ssDNA), (iv) 60-bp poly(rA):poly(dT) (hybrid), or (v) 60-bp poly(dA):poly(dT) (dsDNA). Arrow indicates the expected peak for cGAMP (2′–5′), the product of cGAS catalyzed reaction.

A–D HADDOCK models of different double-stranded nucleic acids in the crystal structure of pig cGAS are shown (the cGAS structure is based on the cGAS:dsDNA PDB ID 4KB6). The DNA strand is shown in green, whereas the RNA strand is depicted in pink. (A) dsDNA bound to cGAS (PDB ID 4KB6). (B, C) Best docking results of an RNA:DNA hybrid in two different orientations (PDB ID 4KB6 chain A) into cGAS are depicted. Hybrid molecules have been located in the cleft formed by the Zn-thumb and Arg150 of cGAS. (D) The best docking solution of dsRNA into the dsDNA binding region of cGAS.
E, F Cartoon and surface representation of superposition of dsDNA (blue), DNA:RNA hybrid (green) and dsRNA (yellow) molecules based on published structures.
References
-
- Ablasser A, Hertrich C, Wassermann R, Hornung V. Nucleic acid driven sterile inflammation. Clin Immunol. 2013b;147:207–215. - PubMed
-
- Ablasser A, Hemmerling I, Schmid-Burgk JL, Behrendt R, Roers A, Hornung V. TREX1 deficiency triggers cell-autonomous immunity in a cGAS-dependent manner. J Immunol. 2014;192:5993–5997. - PubMed
-
- Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

