Salicylic acid modulates levels of phosphoinositide dependent-phospholipase C substrates and products to remodel the Arabidopsis suspension cell transcriptome
- PMID: 25426125
- PMCID: PMC4227474
- DOI: 10.3389/fpls.2014.00608
Salicylic acid modulates levels of phosphoinositide dependent-phospholipase C substrates and products to remodel the Arabidopsis suspension cell transcriptome
Erratum in
-
Corrigendum: Salicylic acid modulates levels of phosphoinositide dependent-phospholipase C substrates and products to remodel the Arabidopsis suspension cell transcriptome.Front Plant Sci. 2016 Jan 28;7:36. doi: 10.3389/fpls.2016.00036. eCollection 2016. Front Plant Sci. 2016. PMID: 26858744 Free PMC article.
Abstract
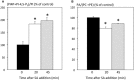
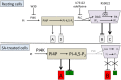
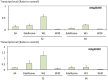
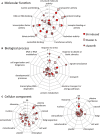
Basal phosphoinositide-dependent phospholipase C (PI-PLC) activity controls gene expression in Arabidopsis suspension cells and seedlings. PI-PLC catalyzes the production of phosphorylated inositol and diacylglycerol (DAG) from phosphoinositides. It is not known how PI-PLC regulates the transcriptome although the action of DAG-kinase (DGK) on DAG immediately downstream from PI-PLC is responsible for some of the regulation. We previously established a list of genes whose expression is affected in the presence of PI-PLC inhibitors. Here this list of genes was used as a signature in similarity searches of curated plant hormone response transcriptome data. The strongest correlations obtained with the inhibited PI-PLC signature were with salicylic acid (SA) treatments. We confirm here that in Arabidopsis suspension cells SA treatment leads to an increase in phosphoinositides, then demonstrate that SA leads to a significant 20% decrease in phosphatidic acid, indicative of a decrease in PI-PLC products. Previous sets of microarray data were re-assessed. The SA response of one set of genes was dependent on phosphoinositides. Alterations in the levels of a second set of genes, mostly SA-repressed genes, could be related to decreases in PI-PLC products that occur in response to SA action. Together, the two groups of genes comprise at least 40% of all SA-responsive genes. Overall these two groups of genes are distinct in the functional categories of the proteins they encode, their promoter cis-elements and their regulation by DGK or phospholipase D. SA-regulated genes dependent on phosphoinositides are typical SA response genes while those with an SA response that is possibly dependent on PI-PLC products are less SA-specific. We propose a model in which SA inhibits PI-PLC activity and alters levels of PI-PLC products and substrates, thereby regulating gene expression divergently.
Keywords: Arabidopsis; diacylglycerol kinase; hormone transduction; lipid signaling; phospholipase C; salicylic acid; trancriptomic.
Figures




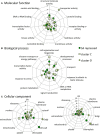

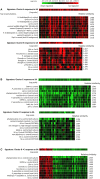


References
-
- Altuzar-Molina A. R., Armando Munoz-Sanchez J., Vazquez-Flota F., Monforte-Gonzalez M., Racagni-Di Palma G., Teresa Hernandez-Sotomayor S. M. (2011). Phospholipidic signaling and vanillin production in response to salicylic acid and methyl jasmonate in Capsicum chinense. Plant Physiol. Biochem. 49, 151–158. 10.1016/j.plaphy.2010.11.005 - DOI - PubMed
-
- Boggs K. P., Rock C. O., Jackowski S. (1995). Lysophosphatidylcholine and 1-O-octadecyl-2-O-methyl-rac-glycero-3-phosphocholine inhibit the CDP-choline pathway of phosphatidylcholine synthesis at the CTP:phosphocholine cytidylyltransferase step. J. Biol. Chem. 270, 7757–7764. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

