Reducing INDEL calling errors in whole genome and exome sequencing data
- PMID: 25426171
- PMCID: PMC4240813
- DOI: 10.1186/s13073-014-0089-z
Reducing INDEL calling errors in whole genome and exome sequencing data
Abstract
Background: INDELs, especially those disrupting protein-coding regions of the genome, have been strongly associated with human diseases. However, there are still many errors with INDEL variant calling, driven by library preparation, sequencing biases, and algorithm artifacts.
Methods: We characterized whole genome sequencing (WGS), whole exome sequencing (WES), and PCR-free sequencing data from the same samples to investigate the sources of INDEL errors. We also developed a classification scheme based on the coverage and composition to rank high and low quality INDEL calls. We performed a large-scale validation experiment on 600 loci, and find high-quality INDELs to have a substantially lower error rate than low-quality INDELs (7% vs. 51%).
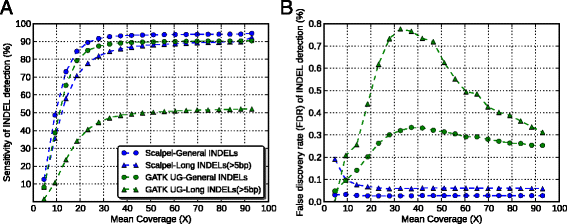
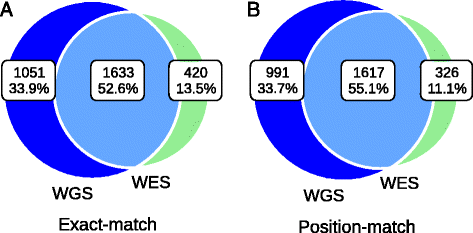
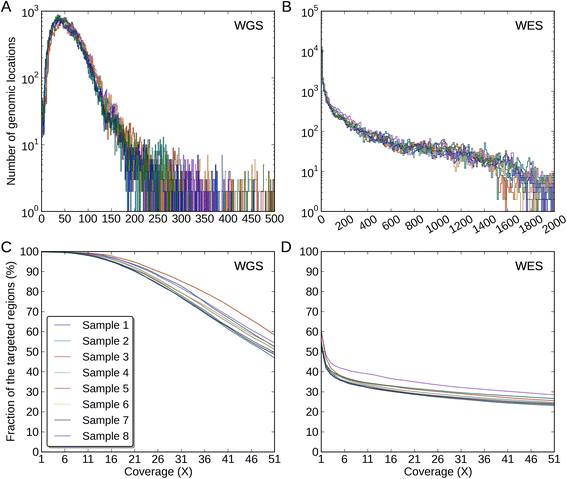
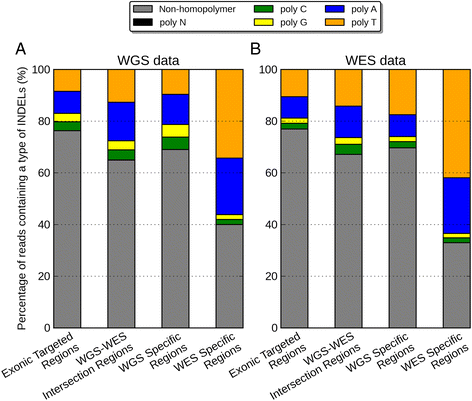
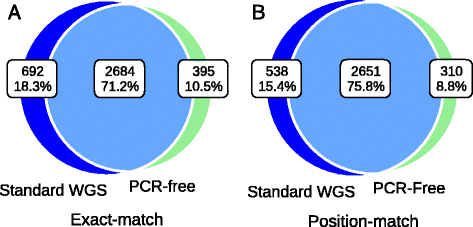
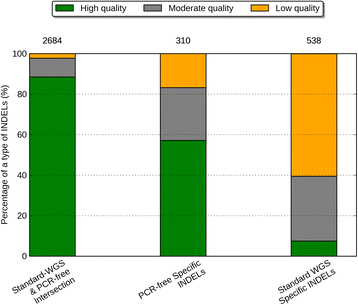
Results: Simulation and experimental data show that assembly based callers are significantly more sensitive and robust for detecting large INDELs (>5 bp) than alignment based callers, consistent with published data. The concordance of INDEL detection between WGS and WES is low (53%), and WGS data uniquely identifies 10.8-fold more high-quality INDELs. The validation rate for WGS-specific INDELs is also much higher than that for WES-specific INDELs (84% vs. 57%), and WES misses many large INDELs. In addition, the concordance for INDEL detection between standard WGS and PCR-free sequencing is 71%, and standard WGS data uniquely identifies 6.3-fold more low-quality INDELs. Furthermore, accurate detection with Scalpel of heterozygous INDELs requires 1.2-fold higher coverage than that for homozygous INDELs. Lastly, homopolymer A/T INDELs are a major source of low-quality INDEL calls, and they are highly enriched in the WES data.
Conclusions: Overall, we show that accuracy of INDEL detection with WGS is much greater than WES even in the targeted region. We calculated that 60X WGS depth of coverage from the HiSeq platform is needed to recover 95% of INDELs detected by Scalpel. While this is higher than current sequencing practice, the deeper coverage may save total project costs because of the greater accuracy and sensitivity. Finally, we investigate sources of INDEL errors (for example, capture deficiency, PCR amplification, homopolymers) with various data that will serve as a guideline to effectively reduce INDEL errors in genome sequencing.
Figures






References
-
- Gudmundsson J, Sulem P, Gudbjartsson DF, Masson G, Agnarsson BA, Benediktsdottir KR, Sigurdsson A, Magnusson OT, Gudjonsson SA, Magnusdottir DN, Johannsdottir H, Helgadottir HT, Stacey SN, Jonasdottir N, Olafsdottir SB, Thorleifsson G, Jonasson JG, Tryggvadottir L, Navarrete S, Fuertes F, Helfand BT, Hu Q, Csiki IE, Mates IN, Jinga V, Aben KKH, van Oort IM, Vermeulen SH, Donovan JL, Hamdy FC, et al. A study based on whole-genome sequencing yields a rare variant at 8q24 associated with prostate cancer. Nat Genet. 2012;44:1326–1329. doi: 10.1038/ng.2437. - DOI - PMC - PubMed
-
- Rope AF, Wang K, Evjenth R, Xing J, Johnston JJ, Swensen JJ, Johnson WE, Moore B, Huff CD, Bird LM, Carey JC, Opitz JM, Stevens CA, Jiang T, Schank C, Fain HD, Robison R, Dalley B, Chin S, South ST, Pysher TJ, Jorde LB, Hakonarson H, Lillehaug JR, Biesecker LG, Yandell M, Arnesen T, Lyon GJ. Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. Am J Hum Genet. 2011;89:28–43. doi: 10.1016/j.ajhg.2011.05.017. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

