Inhibition of autophagy as a new means of improving chemotherapy efficiency in high-LC3B triple-negative breast cancers
- PMID: 25427136
- PMCID: PMC4502743
- DOI: 10.4161/15548627.2014.981788
Inhibition of autophagy as a new means of improving chemotherapy efficiency in high-LC3B triple-negative breast cancers
Abstract
The triple-negative breast cancer (TN BC) subtype is the most aggressive form of invasive BC. Despite intensive efforts to improve BC treatments, patients with TN BC continue to exhibit poor survival, with half developing resistance to chemotherapy. Here we identify autophagy as a key mechanism in the progression and chemoresistance of a subset of TN tumors. We demonstrate that LC3B, a protein involved in autophagosome formation, is a reliable marker of poor prognosis in TN BC, validating this prognostic value at both the mRNA and protein levels in several independent cohorts. We also show that LC3B has no prognostic value for other BC subtypes (Luminal or HER2 BC), thus revealing a specific impact of autophagy on TN tumors. Autophagy is essential for the proliferative and invasive properties in 3D of TN BC cells characterized by high LC3B levels. Interestingly, the activity of the transcriptional co-activator YAP1 (Yes-associated protein 1) is regulated by the autophagy process and we identify YAP1 as a new actor in the autophagy-dependent proliferative and invasive properties of high-LC3B TN BC. Finally, inhibiting autophagy by silencing ATG5 or ATG7 significantly impaired high-LC3B TN tumor growth in vivo. Moreover, using a patient-derived TN tumor transplanted into mice, we show that an autophagy inhibitor, chloroquine, potentiates the effects of chemotherapeutic agents. Overall, our data identify LC3B as a new prognostic marker for TN BC and the inhibition of autophagy as a promising therapeutic strategy for TN BC patients.
Keywords: 3-dimensional culture; 3D, 3-dimensions; AC, adriamycin and cyclophosphamide; ACTB, actin, β; AP2A1/adaptin, adaptor-related protein complex 2, α 1 subunit; ATG, autophagy-related; BC, breast cancer; BECN1, Beclin 1, autophagy related; BafA1, bafilomycin A1; Ctrl, control; DFS, disease-free survival; EBSS, Earle's balanced salt solution; ERBB2/HER2, v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HScore, histological scoring; IHC, immunohistochemistry; LC3B; Lum, Luminal; MAP1LC3B/LC3B, microtubule-associated protein one light chain 3 β; OS, overall survival; PDX, patient-derived xenografted tumor; TCGA, The Cancer Genome Atlas; TGI, tumor growth inhibition; TN BC, triple-negative breast cancer; YAP1; YAP1, Yes-associated protein 1; autophagy; breast cancers; i.p., intra-peritoneal; prognosis; response to treatment; sem, standard error of mean; three-MA, 3-methyladenine.
Figures

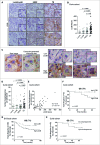
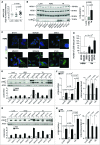

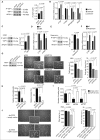


References
-
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin 2010; 60:277-300; PMID:20610543; http://dx.doi.org/10.3322/caac.20073 - DOI - PubMed
-
- Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JWW, Comber H, Forman D, Bray F. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer Oxf Engl 1990 2013; 49:1374-403 - PubMed
-
- Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, Ollila DW, Sartor CI, Graham ML, Perou CM. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res Off J Am Assoc Cancer Res 2007; 13:2329-34; http://dx.doi.org/10.1158/1078-0432.CCR-06-1109 - DOI - PubMed
-
- Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med 2010; 363:1938-48; PMID:21067385; http://dx.doi.org/10.1056/NEJMra1001389 - DOI - PubMed
-
- Kennecke H, Yerushalmi R, Woods R, Cheang MCU, Voduc D, Speers CH, Nielsen TO, Gelmon K. Metastatic behavior of breast cancer subtypes. J Clin Oncol Off J Am Soc Clin Oncol 2010; 28:3271-7; http://dx.doi.org/10.1200/JCO.2009.25.9820 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
