Characteristics of emerging human-pathogenic Escherichia coli O26:H11 strains isolated in France between 2010 and 2013 and carrying the stx2d gene only
- PMID: 25428148
- PMCID: PMC4298503
- DOI: 10.1128/JCM.02290-14
Characteristics of emerging human-pathogenic Escherichia coli O26:H11 strains isolated in France between 2010 and 2013 and carrying the stx2d gene only
Abstract
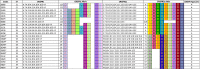
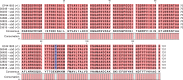
Strains of Escherichia coli O26:H11 that were positive for stx2 alone (n = 23), which were not epidemiologically related or part of an outbreak, were isolated from pediatric patients in France between 2010 and 2013. We were interested in comparing these strains with the new highly virulent stx2a-positive E. coli O26 clone sequence type 29 (ST29) that has emerged recently in Europe, and we tested them by multilocus sequence typing (MLST), stx2 subtyping, clustered regularly interspaced short palindromic repeat (CRISPR) sequencing, and plasmid (ehxA, katP, espP, and etpD) and chromosomal (Z2098, espK, and espV) virulence gene profiling. We showed that 16 of the 23 strains appeared to correspond to this new clone, but the characteristics of 12 strains differed significantly from the previously described characteristics, with negative results for both plasmid and chromosomal genetic markers. These 12 strains exhibited a ST29 genotype and related CRISPR arrays (CRISPR2a alleles 67 or 71), suggesting that they evolved in a common environment. This finding was corroborated by the presence of stx2d in 7 of the 12 ST29 strains. This is the first time that E. coli O26:H11 carrying stx2d has been isolated from humans. This is additional evidence of the continuing evolution of virulent Shiga toxin-producing E. coli (STEC) O26 strains. A new O26:H11 CRISPR PCR assay, SP_O26_E, has been developed for detection of these 12 particular ST29 strains of E. coli O26:H11. This test is useful to better characterize the stx2-positive O26:H11 clinical isolates, which are associated with severe clinical outcomes such as bloody diarrhea and hemolytic uremic syndrome.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Characterization of the Shiga toxin-producing Escherichia coli O26 isolated from human in Poland between 1996 and 2014.Lett Appl Microbiol. 2015 Jun;60(6):605-8. doi: 10.1111/lam.12413. Epub 2015 Apr 8. Lett Appl Microbiol. 2015. PMID: 25758912
-
Genetic Diversity and Pathogenic Potential of Attaching and Effacing Escherichia coli O26:H11 Strains Recovered from Bovine Feces in the United States.Appl Environ Microbiol. 2015 Jun;81(11):3671-8. doi: 10.1128/AEM.00397-15. Epub 2015 Mar 20. Appl Environ Microbiol. 2015. PMID: 25795673 Free PMC article.
-
Large-Scale Phylogenetic Analysis Reveals a New Genetic Clade among Escherichia coli O26 Strains.Microbiol Spectr. 2022 Feb 23;10(1):e0252521. doi: 10.1128/spectrum.02525-21. Epub 2022 Feb 2. Microbiol Spectr. 2022. PMID: 35107330 Free PMC article.
-
Genetic characterization of Shiga toxin-producing Escherichia coli O26:H11 strains isolated from animal, food, and clinical samples.Front Cell Infect Microbiol. 2015 Oct 20;5:74. doi: 10.3389/fcimb.2015.00074. eCollection 2015. Front Cell Infect Microbiol. 2015. PMID: 26539413 Free PMC article.
-
Taxonomy Meets Public Health: The Case of Shiga Toxin-Producing Escherichia coli.Microbiol Spectr. 2014 Jun;2(3). doi: 10.1128/microbiolspec.EHEC-0019-2013. Microbiol Spectr. 2014. PMID: 26103973 Review.
Cited by
-
Whole genome sequencing based typing and characterisation of Shiga-toxin producing Escherichia coli strains belonging to O157 and O26 serotypes and isolated in dairy farms.Ital J Food Saf. 2019 Feb 8;7(4):7673. doi: 10.4081/ijfs.2018.7673. eCollection 2018 Dec 31. Ital J Food Saf. 2019. PMID: 30854339 Free PMC article.
-
Change in the Structure of Escherichia coli Population and the Pattern of Virulence Genes along a Rural Aquatic Continuum.Front Microbiol. 2017 Apr 18;8:609. doi: 10.3389/fmicb.2017.00609. eCollection 2017. Front Microbiol. 2017. PMID: 28458656 Free PMC article.
-
Enhancing detection of STEC in the meat industry: insights into virulence of priority STEC.Front Microbiol. 2025 Feb 12;16:1543686. doi: 10.3389/fmicb.2025.1543686. eCollection 2025. Front Microbiol. 2025. PMID: 40012779 Free PMC article.
-
Heterogeneity in Induction Level, Infection Ability, and Morphology of Shiga Toxin-Encoding Phages (Stx Phages) from Dairy and Human Shiga Toxin-Producing Escherichia coli O26:H11 Isolates.Appl Environ Microbiol. 2016 Jan 29;82(7):2177-2186. doi: 10.1128/AEM.03463-15. Appl Environ Microbiol. 2016. PMID: 26826235 Free PMC article.
-
Revisiting the STEC Testing Approach: Using espK and espV to Make Enterohemorrhagic Escherichia coli (EHEC) Detection More Reliable in Beef.Front Microbiol. 2016 Jan 22;7:1. doi: 10.3389/fmicb.2016.00001. eCollection 2016. Front Microbiol. 2016. PMID: 26834723 Free PMC article.
References
-
- European Food Safety Authority, European Centre for Disease Prevention and Control. 2012. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2010. EFSA J 10:2597. doi:10.2903/j.efsa.2012.2597. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

