OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders
- PMID: 25428349
- PMCID: PMC4383985
- DOI: 10.1093/nar/gku1205
OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders
Abstract
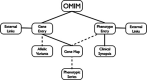
Online Mendelian Inheritance in Man, OMIM(®), is a comprehensive, authoritative and timely research resource of curated descriptions of human genes and phenotypes and the relationships between them. The new official website for OMIM, OMIM.org (http://omim.org), was launched in January 2011. OMIM is based on the published peer-reviewed biomedical literature and is used by overlapping and diverse communities of clinicians, molecular biologists and genome scientists, as well as by students and teachers of these disciplines. Genes and phenotypes are described in separate entries and are given unique, stable six-digit identifiers (MIM numbers). OMIM entries have a structured free-text format that provides the flexibility necessary to describe the complex and nuanced relationships between genes and genetic phenotypes in an efficient manner. OMIM also has a derivative table of genes and genetic phenotypes, the Morbid Map. OMIM.org has enhanced search capabilities such as genome coordinate searching and thesaurus-enhanced search term options. Phenotypic series have been created to facilitate viewing genetic heterogeneity of phenotypes. Clinical synopsis features are enhanced with UMLS, Human Phenotype Ontology and Elements of Morphology terms and image links. All OMIM data are available for FTP download and through an API. MIMmatch is a novel outreach feature to disseminate updates and encourage collaboration.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- McKusick V.A. Mendelian Inheritance in Man: A Catalog of Human Genes and Genetic Disorders. 12th edn. Baltimore: Johns Hopkins University Press; 1998.
-
- Cornet R. Definitions and qualifiers in SNOMED CT. Methods Inf. Med. 2009;48:178–183. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

