The BioGRID interaction database: 2015 update
- PMID: 25428363
- PMCID: PMC4383984
- DOI: 10.1093/nar/gku1204
The BioGRID interaction database: 2015 update
Abstract
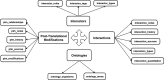
The Biological General Repository for Interaction Datasets (BioGRID: http://thebiogrid.org) is an open access database that houses genetic and protein interactions curated from the primary biomedical literature for all major model organism species and humans. As of September 2014, the BioGRID contains 749,912 interactions as drawn from 43,149 publications that represent 30 model organisms. This interaction count represents a 50% increase compared to our previous 2013 BioGRID update. BioGRID data are freely distributed through partner model organism databases and meta-databases and are directly downloadable in a variety of formats. In addition to general curation of the published literature for the major model species, BioGRID undertakes themed curation projects in areas of particular relevance for biomedical sciences, such as the ubiquitin-proteasome system and various human disease-associated interaction networks. BioGRID curation is coordinated through an Interaction Management System (IMS) that facilitates the compilation interaction records through structured evidence codes, phenotype ontologies, and gene annotation. The BioGRID architecture has been improved in order to support a broader range of interaction and post-translational modification types, to allow the representation of more complex multi-gene/protein interactions, to account for cellular phenotypes through structured ontologies, to expedite curation through semi-automated text-mining approaches, and to enhance curation quality control.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Mouse Genome Sequencing Consortium. Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

