The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis
- PMID: 25428866
- PMCID: PMC4300739
- DOI: 10.1128/JVI.02596-14
The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis
Abstract
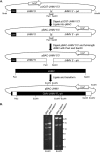
All coronaviruses encode a macrodomain containing ADP-ribose-1"-phosphatase (ADRP) activity within the N terminus of nonstructural protein 3 (nsp3). Previous work showed that mouse hepatitis virus strain A59 (MHV-A59) with a mutated catalytic site (N1348A) replicated similarly to wild-type virus but was unable to cause acute hepatitis in mice. To determine whether this attenuated phenotype is applicable to multiple disease models, we mutated the catalytic residue in the JHM strain of MHV (JHMV), which causes acute and chronic encephalomyelitis, using a newly developed bacterial artificial chromosome (BAC)-based MHV reverse genetics system. Infection of mice with the macrodomain catalytic point mutant virus (N1347A) resulted in reductions in lethality, weight loss, viral titers, proinflammatory cytokine and chemokine expression, and immune cell infiltration in the brain compared to mice infected with wild-type virus. Specifically, macrophages were most affected, with approximately 2.5-fold fewer macrophages at day 5 postinfection in N1347A-infected brains. Tumor necrosis factor (TNF) and interferon (IFN) signaling were not required for effective host control of mutant virus as all N1347A virus-infected mice survived the infection. However, the adaptive immune system was required for protection since N1347A virus was able to cause lethal encephalitis in RAG1(-/-) (recombination activation gene 1 knockout) mice although disease onset was modestly delayed. Overall, these results indicate that the BAC-based MHV reverse genetics system will be useful for studies of JHMV and expand upon previous studies, showing that the macrodomain is critical for the ability of coronaviruses to evade the immune system and promote viral pathogenesis.
Importance: Coronaviruses are an important cause of human and veterinary diseases worldwide. Viral processes that are conserved across a family are likely to be good targets for the development of antiviral therapeutics and vaccines. The macrodomain is a ubiquitous structural domain and is also conserved among all coronaviruses. The coronavirus macrodomain has ADP-ribose-1"-phosphatase activity; however, its function during infection remains unclear as does the reason that coronaviruses have maintained this enzymatic activity throughout evolution. For MHV, this domain has now been shown to promote multiple types of disease, including hepatitis and encephalitis. These data indicate that this domain is vital for the virus to replicate and cause disease. Understanding the mechanism used by this enzyme to promote viral pathogenesis will open up novel avenues for therapies and may give further insight into the role of macrodomain proteins in the host cell since these proteins are found in all living organisms.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures





References
-
- Neuman BW, Joseph JS, Saikatendu KS, Serrano P, Chatterjee A, Johnson MA, Liao L, Klaus JP, Yates JR III, Wuthrich K, Stevens RC, Buchmeier MJ, Kuhn P. 2008. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J Virol 82:5279–5294. doi:10.1128/JVI.02631-07. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

