Receptor recognition mechanisms of coronaviruses: a decade of structural studies
- PMID: 25428871
- PMCID: PMC4338876
- DOI: 10.1128/JVI.02615-14
Receptor recognition mechanisms of coronaviruses: a decade of structural studies
Abstract
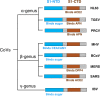
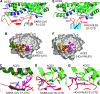
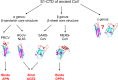
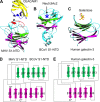
Receptor recognition by viruses is the first and essential step of viral infections of host cells. It is an important determinant of viral host range and cross-species infection and a primary target for antiviral intervention. Coronaviruses recognize a variety of host receptors, infect many hosts, and are health threats to humans and animals. The receptor-binding S1 subunit of coronavirus spike proteins contains two distinctive domains, the N-terminal domain (S1-NTD) and the C-terminal domain (S1-CTD), both of which can function as receptor-binding domains (RBDs). S1-NTDs and S1-CTDs from three major coronavirus genera recognize at least four protein receptors and three sugar receptors and demonstrate a complex receptor recognition pattern. For example, highly similar coronavirus S1-CTDs within the same genus can recognize different receptors, whereas very different coronavirus S1-CTDs from different genera can recognize the same receptor. Moreover, coronavirus S1-NTDs can recognize either protein or sugar receptors. Structural studies in the past decade have elucidated many of the puzzles associated with coronavirus-receptor interactions. This article reviews the latest knowledge on the receptor recognition mechanisms of coronaviruses and discusses how coronaviruses have evolved their complex receptor recognition pattern. It also summarizes important principles that govern receptor recognition by viruses in general.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures




References
-
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong SX, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 348:1953–1966. doi: 10.1056/NEJMoa030781. - DOI - PubMed
-
- Peiris JSM, Lai ST, Poon LLM, Guan Y, Yam LYC, Lim W, Nicholls J, Yee WKS, Yan WW, Cheung MT, Cheng VCC, Chan KH, Tsang DNC, Yung RWH, Ng TK, Yuen KY. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

