Plant-polysaccharide-degrading enzymes from Basidiomycetes
- PMID: 25428937
- PMCID: PMC4248655
- DOI: 10.1128/MMBR.00035-14
Plant-polysaccharide-degrading enzymes from Basidiomycetes
Abstract
Basidiomycete fungi subsist on various types of plant material in diverse environments, from living and dead trees and forest litter to crops and grasses and to decaying plant matter in soils. Due to the variation in their natural carbon sources, basidiomycetes have highly varied plant-polysaccharide-degrading capabilities. This topic is not as well studied for basidiomycetes as for ascomycete fungi, which are the main sources of knowledge on fungal plant polysaccharide degradation. Research on plant-biomass-decaying fungi has focused on isolating enzymes for current and future applications, such as for the production of fuels, the food industry, and waste treatment. More recently, genomic studies of basidiomycete fungi have provided a profound view of the plant-biomass-degrading potential of wood-rotting, litter-decomposing, plant-pathogenic, and ectomycorrhizal (ECM) basidiomycetes. This review summarizes the current knowledge on plant polysaccharide depolymerization by basidiomycete species from diverse habitats. In addition, these data are compared to those for the most broadly studied ascomycete genus, Aspergillus, to provide insight into specific features of basidiomycetes with respect to plant polysaccharide degradation.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures
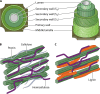







References
-
- Carlile MJ, Watkinson SC, Gooday GW. 2001. The fungi, 2nd ed. Academic Press, London, United Kingdom.
-
- Deacon JW. 2005. Fungal biology. Blackwell, Oxford, United Kingdom.
-
- Eriksson K, Blanchette RA, Ander P. 1990. Microbial and enzymatic degradation of wood and wood components. Springer-Verlag, Berlin, Germany.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

