The formin FMNL3 assembles plasma membrane protrusions that participate in cell-cell adhesion
- PMID: 25428984
- PMCID: PMC4310738
- DOI: 10.1091/mbc.E14-07-1247
The formin FMNL3 assembles plasma membrane protrusions that participate in cell-cell adhesion
Abstract
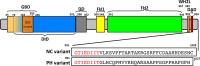
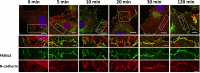
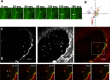
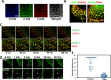
FMNL3 is a vertebrate-specific formin protein previously shown to play a role in angiogenesis and cell migration. Here we define the cellular localization of endogenous FMNL3, the dynamics of GFP-tagged FMNL3 during cell migration, and the effects of FMNL3 suppression in mammalian culture cells. The majority of FMNL3 localizes in a punctate pattern, with >95% of these puncta being indistinguishable from the plasma membrane by fluorescence microscopy. A small number of dynamic cytoplasmic FMNL3 patches also exist, which enrich near cell-cell contact sites and fuse with the plasma membrane at these sites. These cytoplasmic puncta appear to be part of larger membranes of endocytic origin. On the plasma membrane, FMNL3 enriches particularly in filopodia and membrane ruffles and at nascent cell-cell adhesions. FMNL3-containing filopodia occur both at the cell-substratum interface and at cell-cell contacts, with the latter being 10-fold more stable. FMNL3 suppression by siRNA has two major effects: decrease in filopodia and compromised cell-cell adhesion in cells migrating as a sheet. Overall our results suggest that FMNL3 functions in assembly of actin-based protrusions that are specialized for cell-cell adhesion.
© 2015 Gauvin et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures





References
-
- Brevier J, Montero D, Svitkina T, Riveline D. The asymmetric self-assembly mechanism of adherens junctions: a cellular push-pull unit. Phys Biol. 2008;5:016005. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

