The evolution and adaptive potential of transcriptional variation in sticklebacks--signatures of selection and widespread heritability
- PMID: 25429004
- PMCID: PMC4327155
- DOI: 10.1093/molbev/msu328
The evolution and adaptive potential of transcriptional variation in sticklebacks--signatures of selection and widespread heritability
Abstract
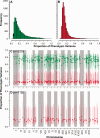
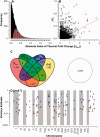
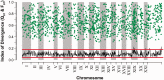
Evidence implicating differential gene expression as a significant driver of evolutionary novelty continues to accumulate, but our understanding of the underlying sources of variation in expression, both environmental and genetic, is wanting. Heritability in particular may be underestimated when inferred from genetic mapping studies, the predominant "genetical genomics" approach to the study of expression variation. Such uncertainty represents a fundamental limitation to testing for adaptive evolution at the transcriptomic level. By studying the inheritance of expression levels in 10,495 genes (10,527 splice variants) in a threespine stickleback pedigree consisting of 563 individuals, half of which were subjected to a thermal treatment, we show that 74-98% of transcripts exhibit significant additive genetic variance. Dominance variance is also prevalent (41-99% of transcripts), and genetic sources of variation seem to play a more significant role in expression variance in the liver than a key environmental variable, temperature. Among-population comparisons suggest that the majority of differential expression in the liver is likely due to neutral divergence; however, we also show that signatures of directional selection may be more prevalent than those of stabilizing selection. This predominantly aligns with the neutral model of evolution for gene expression but also suggests that natural selection may still act on transcriptional variation in the wild. As genetic variation both within- and among-populations ultimately defines adaptive potential, these results indicate that broad adaptive potential may be found within the transcriptome.
Keywords: Gasterosteus aculeatus; QST; gene expression; microarray; quantitative genetics; transcriptome.
© The Author 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
References
-
- Aubin-Horth N, Renn SCP. Genomic reaction norms: using integrative biology to understand molecular mechanisms of phenotypic plasticity. Mol Ecol. 2009;18:3763–3780. - PubMed
-
- Bar-Even A, Paulsson J, Maheshri N, Carmi M, O’Shea E, Pilpel Y, Barkai N. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–643. - PubMed
-
- Barber I, Arnott SA. Split-clutch IVF: a technique to examine indirect fitness consequences of mate preferences in sticklebacks. Behaviour. 2000;137:1129–1140.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

