InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic
- PMID: 25429972
- PMCID: PMC4383983
- DOI: 10.1093/nar/gku1203
InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic
Abstract
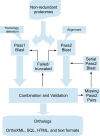
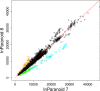
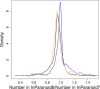
The InParanoid database (http://InParanoid.sbc.su.se) provides a user interface to orthologs inferred by the InParanoid algorithm. As there are now international efforts to curate and standardize complete proteomes, we have switched to using these resources rather than gathering and curating the proteomes ourselves. InParanoid release 8 is based on the 66 reference proteomes that the 'Quest for Orthologs' community has agreed on using, plus 207 additional proteomes from the UniProt complete proteomes--in total 273 species. These represent 246 eukaryotes, 20 bacteria and seven archaea. Compared to the previous release, this increases the number of species by 173% and the number of pairwise species comparisons by 650%. In turn, the number of ortholog groups has increased by 423%. We present the contents and usages of InParanoid 8, and a detailed analysis of how the proteome content has changed since the previous release.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Fitch W.M. Distinguishing homologous from analogous proteins. Syst. Zoolog. 1970;19:99–113. - PubMed
-
- Sonnhammer E.L., Koonin E.V. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002;18:619–620. - PubMed
-
- Remm M., Storm C.E., Sonnhammer E.L. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

