Effects of the deletion and over-expression of Fusarium graminearum gene FgHal2 on host response to mycovirus Fusarium graminearum virus 1
- PMID: 25431083
- PMCID: PMC6638490
- DOI: 10.1111/mpp.12221
Effects of the deletion and over-expression of Fusarium graminearum gene FgHal2 on host response to mycovirus Fusarium graminearum virus 1
Abstract
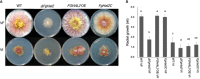
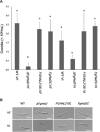
The mycovirus Fusarium graminearum virus 1 (FgV1) is associated with reduced virulence (hypovirulence) of Fusarium graminearum. Transcriptomic and proteomic expression profiling have shown that many F. graminearum genes are differentially expressed as a consequence of FgV1 infection. Several of these genes may be related to the maintenance of the virus life cycle. The host gene, FgHal2, which has a highly conserved 3'-phosphoadenosine 5'-phosphatase (PAP phosphatase-like) domain or inositol monophosphatase (IMPase) superfamily domain, shows reduced expression in response to FgV1 infection. We generated targeted gene deletion and over-expression mutants to clarify the possible function(s) of FgHal2 and its relationship to FgV1. The gene deletion mutant showed retarded growth, reduced aerial mycelia formation and reduced pigmentation, whereas over-expression mutants were morphologically similar to the wild-type (WT). Furthermore, compared with the WT, the gene deletion mutant produced fewer conidia and these showed abnormal morphology. The FgHal2 expression level was decreased by FgV1 infection at 120 h post-inoculation (hpi), whereas the levels were nine-fold greater for both the virus-free and virus-infected over-expression mutant than for the WT. FgV1 RNA accumulation was decreased in the deletion mutant at 48, 72 and 120 hpi. FgV1 RNA accumulation in the over-expression mutant was reduced relative to that of the WT at 48 and 120 hpi, but was similar to that of the WT at 72 hpi. The vertical transmission rate of FgV1 in the gene deletion mutant was low, suggesting that FgHal2 may be required for the maintenance of FgV1 in the host cell. Together, these results indicate that the putative 3'(2'),5'-bisphosphate nucleotidase gene, FgHal2, has diverse biological functions in the host fungus and may affect the viral RNA accumulation and transmission of FgV1.
Keywords: 3′(2′); 5′-bisphosphate nucleotidase; FgHal2; Fusarium graminearum; Fusarium graminearum virus 1; secondary metabolism; virus-host interaction.
© 2014 BSPP AND JOHN WILEY & SONS LTD.
Figures


Similar articles
-
Mycoviruses in Fusarium Species: An Update.Front Cell Infect Microbiol. 2019 Jul 18;9:257. doi: 10.3389/fcimb.2019.00257. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31380300 Free PMC article. Review.
-
Differential Contribution of RNA Interference Components in Response to Distinct Fusarium graminearum Virus Infections.J Virol. 2018 Apr 13;92(9):e01756-17. doi: 10.1128/JVI.01756-17. Print 2018 May 1. J Virol. 2018. PMID: 29437977 Free PMC article.
-
Expression of a Structural Protein of the Mycovirus FgV-ch9 Negatively Affects the Transcript Level of a Novel Symptom Alleviation Factor and Causes Virus Infection-Like Symptoms in Fusarium graminearum.J Virol. 2018 Aug 16;92(17):e00326-18. doi: 10.1128/JVI.00326-18. Print 2018 Sep 1. J Virol. 2018. PMID: 29899100 Free PMC article.
-
The HEX1 gene of Fusarium graminearum is required for fungal asexual reproduction and pathogenesis and for efficient viral RNA accumulation of Fusarium graminearum virus 1.J Virol. 2013 Sep;87(18):10356-67. doi: 10.1128/JVI.01026-13. Epub 2013 Jul 17. J Virol. 2013. PMID: 23864619 Free PMC article.
-
Exploration of the interactions between mycoviruses and Fusarium graminearum.Adv Virus Res. 2020;106:123-144. doi: 10.1016/bs.aivir.2020.01.004. Epub 2020 Feb 5. Adv Virus Res. 2020. PMID: 32327146 Review.
Cited by
-
A Phenome-Wide Association Study of the Effects of Fusarium graminearum Transcription Factors on Fusarium Graminearum Virus 1 Infection.Front Microbiol. 2021 Feb 11;12:622261. doi: 10.3389/fmicb.2021.622261. eCollection 2021. Front Microbiol. 2021. PMID: 33643250 Free PMC article.
-
Five Questions about Mycoviruses.PLoS Pathog. 2015 Nov 5;11(11):e1005172. doi: 10.1371/journal.ppat.1005172. eCollection 2015. PLoS Pathog. 2015. PMID: 26539725 Free PMC article. Review. No abstract available.
-
Exploring the interaction between endornavirus and Sclerotinia sclerotiorum: mechanisms of phytopathogenic fungal virulence and antivirus.mBio. 2025 Mar 12;16(3):e0336524. doi: 10.1128/mbio.03365-24. Epub 2025 Feb 19. mBio. 2025. PMID: 39969183 Free PMC article.
-
Mycoviruses in Fusarium Species: An Update.Front Cell Infect Microbiol. 2019 Jul 18;9:257. doi: 10.3389/fcimb.2019.00257. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31380300 Free PMC article. Review.
-
The ORF2 protein of Fusarium graminearum virus 1 suppresses the transcription of FgDICER2 and FgAGO1 to limit host antiviral defences.Mol Plant Pathol. 2020 Feb;21(2):230-243. doi: 10.1111/mpp.12895. Epub 2019 Dec 9. Mol Plant Pathol. 2020. PMID: 31815356 Free PMC article.
References
-
- Baek, J.‐H. , Park, J.‐A. , Kim, J.‐M. , Oh, J.‐M. , Park, S.‐M. and Kim, D.‐H. (2014) Functional analysis of a tannic‐acid‐inducible and hypoviral‐regulated small heat‐shock protein Hsp24 from the chestnut blight fungus Cryphonectria parasitica . Mol. Plant–Microbe Interact. 27, 56–65. - PubMed
-
- Belles, J.M. and Serrano, R. (1995) A salt‐sensitive 3′(2),5′‐bisphosphate nucleotidase involved in sulfate activation. Science, 267, 232–234. - PubMed
-
- Chen, H. and Xiong, L. (2010) The bifunctional abiotic stress signalling regulator and endogenous RNA silencing suppressor FIERY1 is required for lateral root formation. Plant Cell Environ. 33, 2180–2190. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

