MetaPSICOV: combining coevolution methods for accurate prediction of contacts and long range hydrogen bonding in proteins
- PMID: 25431331
- PMCID: PMC4382908
- DOI: 10.1093/bioinformatics/btu791
MetaPSICOV: combining coevolution methods for accurate prediction of contacts and long range hydrogen bonding in proteins
Abstract
Motivation: Recent developments of statistical techniques to infer direct evolutionary couplings between residue pairs have rendered covariation-based contact prediction a viable means for accurate 3D modelling of proteins, with no information other than the sequence required. To extend the usefulness of contact prediction, we have designed a new meta-predictor (MetaPSICOV) which combines three distinct approaches for inferring covariation signals from multiple sequence alignments, considers a broad range of other sequence-derived features and, uniquely, a range of metrics which describe both the local and global quality of the input multiple sequence alignment. Finally, we use a two-stage predictor, where the second stage filters the output of the first stage. This two-stage predictor is additionally evaluated on its ability to accurately predict the long range network of hydrogen bonds, including correctly assigning the donor and acceptor residues.
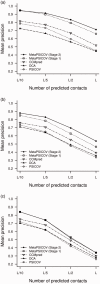
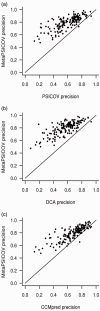
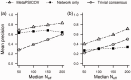
Results: Using the original PSICOV benchmark set of 150 protein families, MetaPSICOV achieves a mean precision of 0.54 for top-L predicted long range contacts-around 60% higher than PSICOV, and around 40% better than CCMpred. In de novo protein structure prediction using FRAGFOLD, MetaPSICOV is able to improve the TM-scores of models by a median of 0.05 compared with PSICOV. Lastly, for predicting long range hydrogen bonding, MetaPSICOV-HB achieves a precision of 0.69 for the top-L/10 hydrogen bonds compared with just 0.26 for the baseline MetaPSICOV.
Availability and implementation: MetaPSICOV is available as a freely available web server at http://bioinf.cs.ucl.ac.uk/MetaPSICOV. Raw data (predicted contact lists and 3D models) and source code can be downloaded from http://bioinf.cs.ucl.ac.uk/downloads/MetaPSICOV.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2014. Published by Oxford University Press.
Figures
Similar articles
-
Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model.PLoS Comput Biol. 2017 Jan 5;13(1):e1005324. doi: 10.1371/journal.pcbi.1005324. eCollection 2017 Jan. PLoS Comput Biol. 2017. PMID: 28056090 Free PMC article.
-
The evolution of contact prediction: evidence that contact selection in statistical contact prediction is changing.Bioinformatics. 2020 Mar 1;36(6):1750-1756. doi: 10.1093/bioinformatics/btz816. Bioinformatics. 2020. PMID: 31693112
-
PSICOV: precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments.Bioinformatics. 2012 Jan 15;28(2):184-90. doi: 10.1093/bioinformatics/btr638. Epub 2011 Nov 17. Bioinformatics. 2012. PMID: 22101153
-
State-of-the-art web services for de novo protein structure prediction.Brief Bioinform. 2021 May 20;22(3):bbaa139. doi: 10.1093/bib/bbaa139. Brief Bioinform. 2021. PMID: 34020540 Review.
-
Prediction of protein contacts from correlated sequence substitutions.Sci Prog. 2013;96(Pt 1):33-42. doi: 10.3184/003685013X13612883013639. Sci Prog. 2013. PMID: 23738436 Free PMC article. Review.
Cited by
-
The PSIPRED Protein Analysis Workbench: 20 years on.Nucleic Acids Res. 2019 Jul 2;47(W1):W402-W407. doi: 10.1093/nar/gkz297. Nucleic Acids Res. 2019. PMID: 31251384 Free PMC article.
-
Machine-designed biotherapeutics: opportunities, feasibility and advantages of deep learning in computational antibody discovery.Brief Bioinform. 2022 Jul 18;23(4):bbac267. doi: 10.1093/bib/bbac267. Brief Bioinform. 2022. PMID: 35830864 Free PMC article. Review.
-
Protein inter-residue contact and distance prediction by coupling complementary coevolution features with deep residual networks in CASP14.Proteins. 2021 Dec;89(12):1911-1921. doi: 10.1002/prot.26211. Epub 2021 Aug 19. Proteins. 2021. PMID: 34382712 Free PMC article.
-
An interactive visualization tool for educational outreach in protein contact map overlap analysis.Front Bioinform. 2024 Mar 15;4:1358550. doi: 10.3389/fbinf.2024.1358550. eCollection 2024. Front Bioinform. 2024. PMID: 38562910 Free PMC article.
-
Recent Applications of Deep Learning Methods on Evolution- and Contact-Based Protein Structure Prediction.Int J Mol Sci. 2021 Jun 2;22(11):6032. doi: 10.3390/ijms22116032. Int J Mol Sci. 2021. PMID: 34199677 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

