Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection
- PMID: 25431490
- PMCID: PMC4409937
- DOI: 10.1126/science.1258025
Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection
Abstract
The capacity of human norovirus (NoV), which causes >90% of global epidemic nonbacterial gastroenteritis, to infect a subset of people persistently may contribute to its spread. How such enteric viruses establish persistent infections is not well understood. We found that antibiotics prevented persistent murine norovirus (MNoV) infection, an effect that was reversed by replenishment of the bacterial microbiota. Antibiotics did not prevent tissue infection or affect systemic viral replication but acted specifically in the intestine. The receptor for the antiviral cytokine interferon-λ, Ifnlr1, as well as the transcription factors Stat1 and Irf3, were required for antibiotics to prevent viral persistence. Thus, the bacterial microbiome fosters enteric viral persistence in a manner counteracted by specific components of the innate immune system.
Copyright © 2015, American Association for the Advancement of Science.
Figures
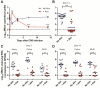
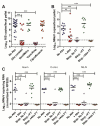
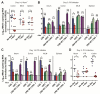
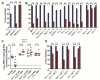
Comment in
-
Immunology. Interfering with interferons.Science. 2015 Jan 16;347(6219):233-4. doi: 10.1126/science.aaa5056. Science. 2015. PMID: 25593173 No abstract available.
References
-
- Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009 Jul 10;138:30. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

