Mosquito genomics. Extensive introgression in a malaria vector species complex revealed by phylogenomics
- PMID: 25431491
- PMCID: PMC4380269
- DOI: 10.1126/science.1258524
Mosquito genomics. Extensive introgression in a malaria vector species complex revealed by phylogenomics
Abstract
Introgressive hybridization is now recognized as a widespread phenomenon, but its role in evolution remains contested. Here, we use newly available reference genome assemblies to investigate phylogenetic relationships and introgression in a medically important group of Afrotropical mosquito sibling species. We have identified the correct species branching order to resolve a contentious phylogeny and show that lineages leading to the principal vectors of human malaria were among the first to split. Pervasive autosomal introgression between these malaria vectors means that only a small fraction of the genome, mainly on the X chromosome, has not crossed species boundaries. Our results suggest that traits enhancing vectorial capacity may be gained through interspecific gene flow, including between nonsister species.
Copyright © 2015, American Association for the Advancement of Science.
Figures
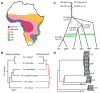
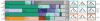
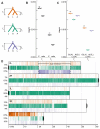

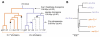
Comment in
-
Evolutionary genomics. Conundrum of jumbled mosquito genomes.Science. 2015 Jan 2;347(6217):27-8. doi: 10.1126/science.aaa3600. Science. 2015. PMID: 25554775 No abstract available.
References
-
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, Hansen NF, Durand EY, Malaspinas AS, Jensen JD, Marques-Bonet T, Alkan C, Prufer K, Meyer M, Burbano HA, Good JM, Schultz R, Aximu-Petri A, Butthof A, Hober B, Hoffner B, Siegemund M, Weihmann A, Nusbaum C, Lander ES, Russ C, Novod N, Affourtit J, Egholm M, Verna C, Rudan P, Brajkovic D, Kucan Z, Gusic I, Doronichev VB, Golovanova LV, Lalueza-Fox C, de la Rasilla M, Fortea J, Rosas A, Schmitz RW, Johnson PL, Eichler EE, Falush D, Birney E, Mullikin JC, Slatkin M, Nielsen R, Kelso J, Lachmann M, Reich D, Paabo S. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. - PMC - PubMed
-
- Dasmahapatra KK, Walters JR, Briscoe AD, Davey JW, Whibley A, Nadeau NJ, Zimin AV, Hughes DS, Ferguson LC, Martin SH, Salazar C, Lewis JJ, Adler S, Ahn SJ, Baker DA, Baxter SW, Chamberlain NL, Chauhan R, Counterman BA, Dalmay T, Gilbert LE, Gordon K, Heckel DG, Hines HM, Hoff KJ, Holland PW, Jacquin-Joly E, Jiggins FM, Jones RT, Kapan DD, Kersey P, Lamas G, Lawson D, Mapleson D, Maroja LS, Martin A, Moxon S, Palmer WJ, Papa R, Papanicolaou A, Pauchet Y, Ray DA, Rosser N, Salzberg SL, Supple MA, Surridge A, Tenger-Trolander A, Vogel H, Wilkinson PA, Wilson D, Yorke JA, Yuan F, Balmuth AL, Eland C, Gharbi K, Thomson M, Gibbs RA, Han Y, Jayaseelan JC, Kovar C, Mathew T, Muzny DM, Ongeri F, Pu LL, Qu J, Thornton RL, Worley KC, Wu YQ, Linares M, Blaxter ML, ffrench-Constant RH, Joron M, Kronforst MR, Mullen SP, Reed RD, Scherer SE, Richards S, Mallet J, McMillan W, Jiggins CD. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487:94–98. - PMC - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
- R01AI085079/AI/NIAID NIH HHS/United States
- R21 AI099528/AI/NIAID NIH HHS/United States
- G1100339/MRC_/Medical Research Council/United Kingdom
- R01AI076584/AI/NIAID NIH HHS/United States
- T32 GM007757/GM/NIGMS NIH HHS/United States
- R21AI101459/AI/NIAID NIH HHS/United States
- R01 AI063508/AI/NIAID NIH HHS/United States
- HHSN272200900039C/AI/NIAID NIH HHS/United States
- R01 AI076584/AI/NIAID NIH HHS/United States
- R21AI099528/AI/NIAID NIH HHS/United States
- R01 AI104956/AI/NIAID NIH HHS/United States
- R01AI104956/AI/NIAID NIH HHS/United States
- R21 AI101459/AI/NIAID NIH HHS/United States
- R01 AI085079/AI/NIAID NIH HHS/United States
- T32GM007757/GM/NIGMS NIH HHS/United States
- R21 AI094289/AI/NIAID NIH HHS/United States
- R21AI094289/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

