MALDI imaging mass spectrometry of N-linked glycans on formalin-fixed paraffin-embedded murine kidney
- PMID: 25434632
- PMCID: PMC4357650
- DOI: 10.1007/s00216-014-8293-7
MALDI imaging mass spectrometry of N-linked glycans on formalin-fixed paraffin-embedded murine kidney
Abstract
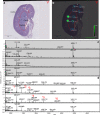
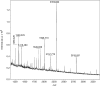
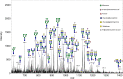
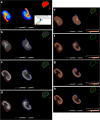
Recent developments in spatial proteomics have paved the way for retrospective in situ mass spectrometry (MS) analyses of formalin-fixed paraffin-embedded clinical tissue samples. This type of analysis is commonly referred to as matrix-assisted laser desorption/ionization (MALDI) imaging. Recently, formalin-fixed paraffin-embedded MALDI imaging analyses were augmented to allow in situ analyses of tissue-specific N-glycosylation profiles. In the present study, we outline an improved automated sample preparation method for N-glycan MALDI imaging, which uses in situ PNGase F-mediated release and measurement of N-linked glycans from sections of formalin-fixed murine kidney. The sum of the presented data indicated that N-glycans can be cleaved from proteins within formalin-fixed tissue and characterized using three strategies: (i) extraction and composition analysis through on-target MALDI MS and liquid chromatography coupled to electrospray ionization ion trap MS; (ii) MALDI profiling, where N-glycans are released and measured from large droplet arrays in situ; and (iii) MALDI imaging, which maps the tissue specificity of N-glycans at a higher resolution. Thus, we present a complete, straightforward method that combines MALDI imaging and characterization of tissue-specific N-glycans and complements existing strategies.
Figures


References
-
- Kam RKT, Poon TCW. The potentials of glycomics in biomarker discovery. Clin Proteom. 2008;4:67–79. doi: 10.1007/s12014-008-9017-9. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

