A chemo-centric view of human health and disease
- PMID: 25435099
- PMCID: PMC4338530
- DOI: 10.1038/ncomms6676
A chemo-centric view of human health and disease
Abstract
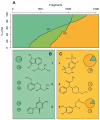
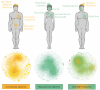
Efforts to compile the phenotypic effects of drugs and environmental chemicals offer the opportunity to adopt a chemo-centric view of human health that does not require detailed mechanistic information. Here we consider thousands of chemicals and analyse the relationship of their structures with adverse and therapeutic responses. Our study includes molecules related to the aetiology of 934 health-threatening conditions and used to treat 835 diseases. We first identify chemical moieties that could be independently associated with each phenotypic effect. Using these fragments, we build accurate predictors for approximately 400 clinical phenotypes, finding many privileged and liable structures. Finally, we connect two diseases if they relate to similar chemical structures. The resulting networks of human conditions are able to predict disease comorbidities, as well as identifying potential drug side effects and opportunities for drug repositioning, and show a remarkable coincidence with clinical observations.
Figures




References
-
- Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug-target network. Nature biotechnology. 2007;25:1119–1126. doi:10.1038/nbt1338. - PubMed
-
- Pujol A, Mosca R, Farres J, Aloy P. Unveiling the role of network and systems biology in drug discovery. Trends in pharmacological sciences. 2010;31:115–123. doi:10.1016/j.tips.2009.11.006. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

