High dimensional biological data retrieval optimization with NoSQL technology
- PMID: 25435347
- PMCID: PMC4248814
- DOI: 10.1186/1471-2164-15-S8-S3
High dimensional biological data retrieval optimization with NoSQL technology
Abstract
Background: High-throughput transcriptomic data generated by microarray experiments is the most abundant and frequently stored kind of data currently used in translational medicine studies. Although microarray data is supported in data warehouses such as tranSMART, when querying relational databases for hundreds of different patient gene expression records queries are slow due to poor performance. Non-relational data models, such as the key-value model implemented in NoSQL databases, hold promise to be more performant solutions. Our motivation is to improve the performance of the tranSMART data warehouse with a view to supporting Next Generation Sequencing data.
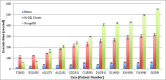
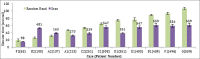
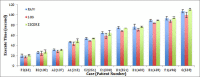
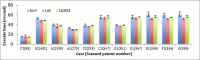
Results: In this paper we introduce a new data model better suited for high-dimensional data storage and querying, optimized for database scalability and performance. We have designed a key-value pair data model to support faster queries over large-scale microarray data and implemented the model using HBase, an implementation of Google's BigTable storage system. An experimental performance comparison was carried out against the traditional relational data model implemented in both MySQL Cluster and MongoDB, using a large publicly available transcriptomic data set taken from NCBI GEO concerning Multiple Myeloma. Our new key-value data model implemented on HBase exhibits an average 5.24-fold increase in high-dimensional biological data query performance compared to the relational model implemented on MySQL Cluster, and an average 6.47-fold increase on query performance on MongoDB.
Conclusions: The performance evaluation found that the new key-value data model, in particular its implementation in HBase, outperforms the relational model currently implemented in tranSMART. We propose that NoSQL technology holds great promise for large-scale data management, in particular for high-dimensional biological data such as that demonstrated in the performance evaluation described in this paper. We aim to use this new data model as a basis for migrating tranSMART's implementation to a more scalable solution for Big Data.
Figures

References
-
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall K a, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database):D991–5. - PMC - PubMed
-
- Kapushesky M, Adamusiak T, Burdett T, Culhane A, Farne A, Filippov A, Holloway E, Klebanov A, Kryvych N, Kurbatova N, Kurnosov P, Malone J, Melnichuk O, Petryszak R, Pultsin N, Rustici G, Tikhonov A, Travillian RS, Williams E, Zorin A, Parkinson H, Brazma A. Gene Expression Atlas update -- a value-added database of microarray and sequencing-based functional genomics experiments. Gene Expr. 2012;40:1–5. - PMC - PubMed
-
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006. pp. 500–501. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

