Large cryptic genomic rearrangements with apparently normal karyotypes detected by array-CGH
- PMID: 25435912
- PMCID: PMC4247713
- DOI: 10.1186/s13039-014-0082-7
Large cryptic genomic rearrangements with apparently normal karyotypes detected by array-CGH
Abstract
Background: Conventional karyotyping (550 bands resolution) is able to identify chromosomal aberrations >5-10 Mb, which represent a known cause of intellectual disability/developmental delay (ID/DD) and/or multiple congenital anomalies (MCA). Array-Comparative Genomic Hybridization (array-CGH) has increased the diagnostic yield of 15-20%.
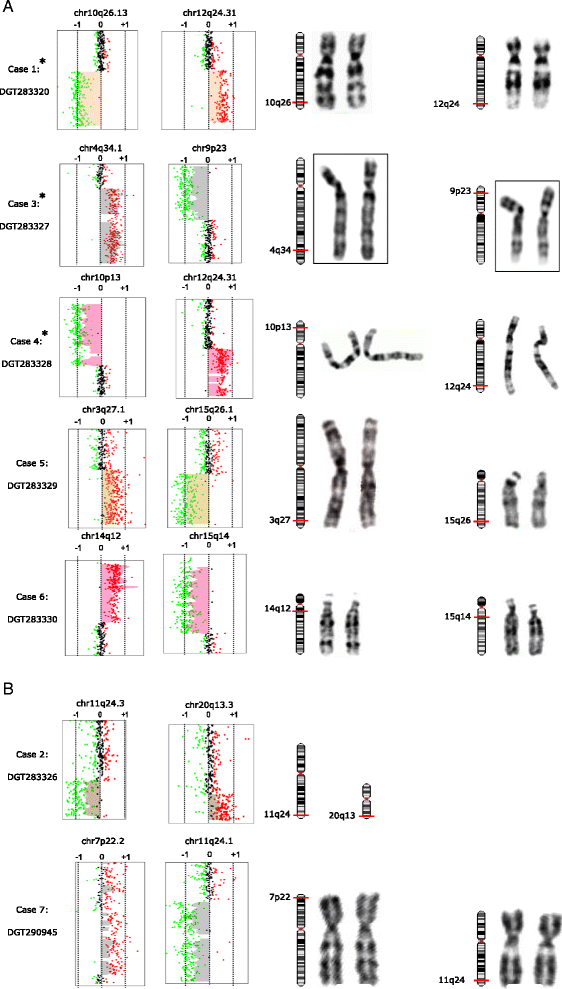
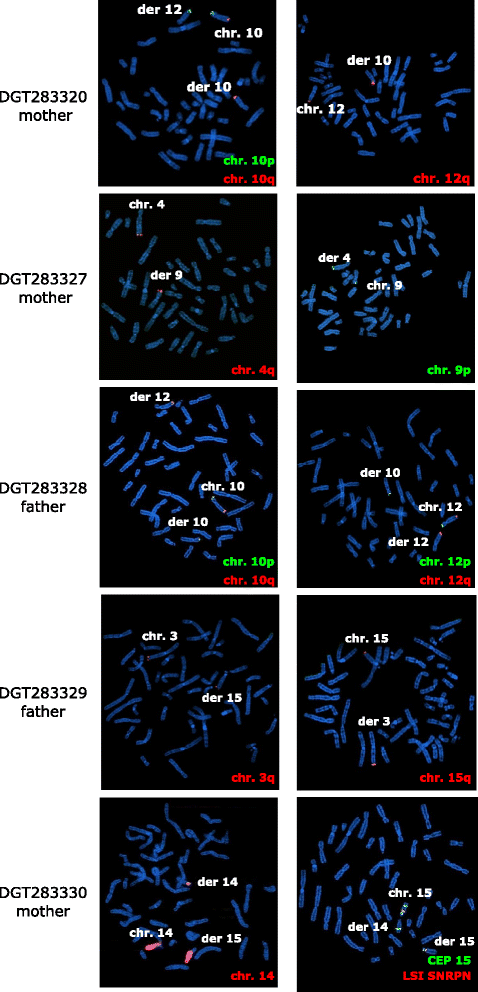
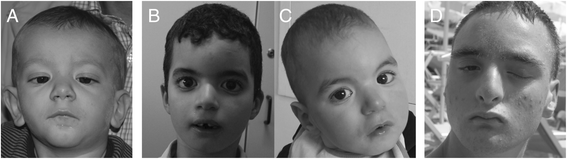
Results: In a cohort of 700 ID/DD cases with or without MCA, including 15 prenatal diagnoses, we identified a subgroup of seven patients with a normal karyotype and a large complex rearrangement detected by array-CGH (at least 6, and up to 18 Mb). FISH analysis could be performed on six cases and showed that rearrangements were translocation derivatives, indistinguishable from a normal karyotype as they involved a similar band pattern and size. Five were inherited from a parent with a balanced translocation, whereas two were apparently de novo. Genes spanning the rearrangements could be associated with some phenotypic features in three cases (case 3: DOCK8; case 4: GATA3, AKR1C4; case 6: AS/PWS deletion, CHRNA7), and in two, likely disease genes were present (case 5: NR2F2, TP63, IGF1R; case 7: CDON). Three of our cases were prenatal diagnoses with an apparently normal karyotype.
Conclusions: Large complex rearrangements of up to 18 Mb, involving chromosomal regions with similar size and band appearance may be overlooked by conventional karyotyping. Array-CGH allows a precise chromosomal diagnosis and recurrence risk definition, further confirming this analysis as a first tier approach to clarify molecular bases of ID/DD and/or MCA. In prenatal tests, array-CGH is confirmed as an important tool to avoid false negative results due to karyotype intrinsic limit of detection.
Keywords: Array-CGH; CNV; GTG-banding; Genomic rearrangement; Intellectual disability; Unbalanced derivative chromosomes.
Figures
References
-
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ, Faucett WA, Feuk L, Friedman JM, Hamosh A, Jackson L, Kaminsky EB, Kok K, Krantz ID, Kuhn RM, Lee C, Ostell JM, Rosenberg C, Scherer SW, Spinner NB, Stavropoulos DJ, Tepperberg JH, Thorland EC, Vermeesch JR, Waggoner DJ, Watson MS, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. - DOI - PMC - PubMed
-
- Shaffer LG, Bejjani BA, Torchia B, Kirkpatrick S, Coppinger J, Ballif BC. The identification of microdeletion syndromes and other chromosome abnormalities: Cytogenetic methods of the past, new technologies for the future. Am J Med Genet C. 2007;145C(4):335–345. doi: 10.1002/ajmg.c.30152. - DOI - PubMed
-
- Lu X, Shaw CA, Patel A, Li J, Cooper ML, Wells WR, Sullivan CM, Sahoo T, Yatsenko SA, Bacino CA, Stankiewicz P, Ou Z, Chinault AC, Beaudet AL, Lupski JR, Cheung SW, Ward PA. Clinical implementation of chromosomal microarray analysis: summary of 2513 postnatal cases. PLoS One. 2007;2:e327. doi: 10.1371/journal.pone.0000327. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

