Coupled protein diffusion and folding in the cell
- PMID: 25436502
- PMCID: PMC4249841
- DOI: 10.1371/journal.pone.0113040
Coupled protein diffusion and folding in the cell
Abstract
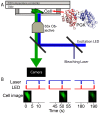
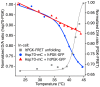
When a protein unfolds in the cell, its diffusion coefficient is affected by its increased hydrodynamic radius and by interactions of exposed hydrophobic residues with the cytoplasmic matrix, including chaperones. We characterize protein diffusion by photobleaching whole cells at a single point, and imaging the concentration change of fluorescent-labeled protein throughout the cell as a function of time. As a folded reference protein we use green fluorescent protein. The resulting region-dependent anomalous diffusion is well characterized by 2-D or 3-D diffusion equations coupled to a clustering algorithm that accounts for position-dependent diffusion. Then we study diffusion of a destabilized mutant of the enzyme phosphoglycerate kinase (PGK) and of its stable control inside the cell. Unlike the green fluorescent protein control's diffusion coefficient, PGK's diffusion coefficient is a non-monotonic function of temperature, signaling 'sticking' of the protein in the cytosol as it begins to unfold. The temperature-dependent increase and subsequent decrease of the PGK diffusion coefficient in the cytosol is greater than a simple size-scaling model suggests. Chaperone binding of the unfolding protein inside the cell is one plausible candidate for even slower diffusion of PGK, and we test the plausibility of this hypothesis experimentally, although we do not rule out other candidates.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

