Proteomic profile and in silico analysis in metastatic melanoma with and without BRAF mutation
- PMID: 25437182
- PMCID: PMC4249853
- DOI: 10.1371/journal.pone.0112025
Proteomic profile and in silico analysis in metastatic melanoma with and without BRAF mutation
Erratum in
-
Correction: Proteomic profile and in silico analysis in metastatic melanoma with and without BRAF mutation.PLoS One. 2015 Mar 30;10(3):e0122943. doi: 10.1371/journal.pone.0122943. eCollection 2015. PLoS One. 2015. PMID: 25822884 Free PMC article. No abstract available.
Abstract
Introduction: Selective inhibitors of BRAF, vemurafenib and dabrafenib are the standard of care for metastatic melanoma patients with BRAF V600, while chemotherapy continued to be widely used in BRAF wild type patients.
Materials and methods: In order to discover novel candidate biomarkers predictive to treatment, serum of 39 metastatic melanoma vemurafenib (n = 19) or chemotherapy (n = 20) treated patients at baseline, at disease control and at progression, were analyzed using SELDI-TOF technology. In silico analysis was used to identify more significant peaks.
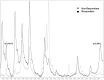
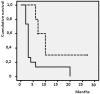
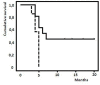
Results: In patients with different BRAF status, we found 5 peptides significantly deregulated, with the down-regulation of the m/z 9176 peak strongly associated with BRAF mutation. At baseline as predictive biomarkers we identified 2 peptides - m/z 6411, 4075 - as significantly up-regulated in responders to chemotherapy and 4 peaks - m/z 5900, 12544, 49124 and 11724 - significantly up-regulated in longer vs shorter responders to vemurafenib. After response, 3 peptides (m/z 4658, 18639, and 9307) resulted significantly down regulated while 3 peptides m/z 9292, 7765 and 9176 appeared up-regulated respectively in chemotherapy and vemurafenib responder patients. In vemurafenib treated patients, 16 peaks appeared deregulated at progression compared to baseline time. In silico analysis identified proteins involved in invasiveness (SLAIN1) and resistance (ABCC12) as well as in the pathway of detoxification (NQO1) and apoptosis (RBM10, TOX3, MTEFD1, TSPO2). Proteins associated with the modulation of neuronal plasticity (RIN1) and regulatory activity factors of gene transcription (KLF17, ZBTB44) were also highlighted.
Conclusion: Our exploratory study highlighted some factors that deserve to be further investigated in order to provide a framework for improving melanoma treatment management through the development of biomarkers which could act as the strongest surrogates of the key biological events in stage IV melanoma.
Conflict of interest statement
Figures
References
-
- Forsea AM, del Marmol V, de Vries E, Bailey EE, Geller AC (2012) Melanoma incidence and mortality in Europe: new estimates, persistent disparities. British Journal of Dermatology 167:1124–30. - PubMed
-
- Eggermont AM, Robert C (2011) New drugs in melanoma: It's a whole new world. European Journal of Cancer 47(14):2150–7. - PubMed
-
- Guida M, Pisconti S, Colucci G (2012) Metastatic Melanoma: the new era of targeted therapy. Expert Opinion on Therapeutic Targets Suppl 2 S61–70. - PubMed
-
- Fecher LA, Cummings SD, Keefe MJ, Alani RM (2007) Toward a Molecular Classification of Melanoma. Journal of Clinical Oncology 25:1606–1620. - PubMed
-
- Robert C, Thomas L, Bondarenko I, O'Day S, Weber J, et al. (2011) Ipilimumab plus Dacarbazine for Previously Untreated Metastatic Melanoma. New England Journal of Medicine 364:2517–2526. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

