CD161 defines a transcriptional and functional phenotype across distinct human T cell lineages
- PMID: 25437561
- PMCID: PMC4250839
- DOI: 10.1016/j.celrep.2014.09.045
CD161 defines a transcriptional and functional phenotype across distinct human T cell lineages
Abstract
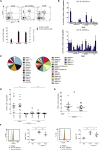
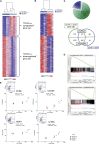
The C-type lectin CD161 is expressed by a large proportion of human T lymphocytes of all lineages, including a population known as mucosal-associated invariant T (MAIT) cells. To understand whether different T cell subsets expressing CD161 have similar properties, we examined these populations in parallel using mass cytometry and mRNA microarray approaches. The analysis identified a conserved CD161++/MAIT cell transcriptional signature enriched in CD161+CD8+ T cells, which can be extended to CD161+ CD4+ and CD161+TCRγδ+ T cells. Furthermore, this led to the identification of a shared innate-like, TCR-independent response to interleukin (IL)-12 plus IL-18 by different CD161-expressing T cell populations. This response was independent of regulation by CD161, which acted as a costimulatory molecule in the context of T cell receptor stimulation. Expression of CD161 hence identifies a transcriptional and functional phenotype, shared across human T lymphocytes and independent of both T cell receptor (TCR) expression and cell lineage.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Aldemir H., Prod’homme V., Dumaurier M.-J., Retiere C., Poupon G., Cazareth J., Bihl F., Braud V.M. Cutting edge: lectin-like transcript 1 is a ligand for the CD161 receptor. J. Immunol. 2005;175:7791–7795. - PubMed
-
- Annibali V., Ristori G., Angelini D.F., Serafini B., Mechelli R., Cannoni S., Romano S., Paolillo A., Abderrahim H., Diamantini A. CD161(high)CD8+T cells bear pathogenetic potential in multiple sclerosis. Brain. 2011;134:542–554. - PubMed
-
- Battistini L., Borsellino G., Sawicki G., Poccia F., Salvetti M., Ristori G., Brosnan C.F. Phenotypic and cytokine analysis of human peripheral blood gamma delta T cells expressing NK cell receptors. J. Immunol. 1997;159:3723–3730. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

