A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease
- PMID: 25437566
- PMCID: PMC4255334
- DOI: 10.1016/j.celrep.2014.09.042
A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease
Abstract
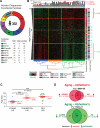
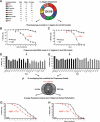
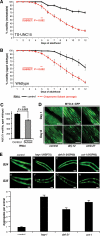
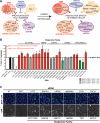
Chaperones are central to the proteostasis network (PN) and safeguard the proteome from misfolding, aggregation, and proteotoxicity. We categorized the human chaperome of 332 genes into network communities using function, localization, interactome, and expression data sets. During human brain aging, expression of 32% of the chaperome, corresponding to ATP-dependent chaperone machines, is repressed, whereas 19.5%, corresponding to ATP-independent chaperones and co-chaperones, are induced. These repression and induction clusters are enhanced in the brains of those with Alzheimer's, Huntington's, or Parkinson's disease. Functional properties of the chaperome were assessed by perturbation in C. elegans and human cell models expressing Aβ, polyglutamine, and Huntingtin. Of 219 C. elegans orthologs, knockdown of 16 enhanced both Aβ and polyQ-associated toxicity. These correspond to 28 human orthologs, of which 52% and 41% are repressed, respectively, in brain aging and disease and 37.5% affected Huntingtin aggregation in human cells. These results identify a critical chaperome subnetwork that functions in aging and disease.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Ahn YY, Bagrow JP, et al. Link communities reveal multiscale complexity in networks. Nature. 2010;466(7307):761–764. - PubMed
-
- Albanese V, Yam AY, et al. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 2006;124(1):75–88. - PubMed
-
- Balch WE, Morimoto RI, et al. Adapting proteostasis for disease intervention. Science. 2008;319(5865):916–919. - PubMed
-
- Behrends C, Langer CA, et al. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Molecular cell. 2006;23(6):887–897. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

