Identifying pathogenicity islands in bacterial pathogenomics using computational approaches
- PMID: 25437607
- PMCID: PMC4235732
- DOI: 10.3390/pathogens3010036
Identifying pathogenicity islands in bacterial pathogenomics using computational approaches
Abstract
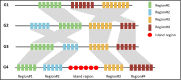
High-throughput sequencing technologies have made it possible to study bacteria through analyzing their genome sequences. For instance, comparative genome sequence analyses can reveal the phenomenon such as gene loss, gene gain, or gene exchange in a genome. By analyzing pathogenic bacterial genomes, we can discover that pathogenic genomic regions in many pathogenic bacteria are horizontally transferred from other bacteria, and these regions are also known as pathogenicity islands (PAIs). PAIs have some detectable properties, such as having different genomic signatures than the rest of the host genomes, and containing mobility genes so that they can be integrated into the host genome. In this review, we will discuss various pathogenicity island-associated features and current computational approaches for the identification of PAIs. Existing pathogenicity island databases and related computational resources will also be discussed, so that researchers may find it to be useful for the studies of bacterial evolution and pathogenicity mechanisms.
Figures

References
-
- Hacker J., Bender L., Ott M., Wingender J., Lund B., Marre R., Goebel W. Deletions of chromosomal regions coding for fimbriae and hemolysins occur in vitro and in vivo in various extraintestinal Escherichia coli isolates. Microb. Pathog. 1990;8:213–225. doi: 10.1016/0882-4010(90)90048-U. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

