Leptospiral pathogenomics
- PMID: 25437801
- PMCID: PMC4243447
- DOI: 10.3390/pathogens3020280
Leptospiral pathogenomics
Abstract
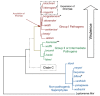
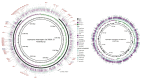
Leptospirosis, caused by pathogenic spirochetes belonging to the genus Leptospira, is a zoonosis with important impacts on human and animal health worldwide. Research on the mechanisms of Leptospira pathogenesis has been hindered due to slow growth of infectious strains, poor transformability, and a paucity of genetic tools. As a result of second generation sequencing technologies, there has been an acceleration of leptospiral genome sequencing efforts in the past decade, which has enabled a concomitant increase in functional genomics analyses of Leptospira pathogenesis. A pathogenomics approach, by coupling of pan-genomic analysis of multiple isolates with sequencing of experimentally attenuated highly pathogenic Leptospira, has resulted in the functional inference of virulence factors. The global Leptospira Genome Project supported by the U.S. National Institute of Allergy and Infectious Diseases to which key scientific contributions have been made from the international leptospirosis research community has provided a new roadmap for comprehensive studies of Leptospira and leptospirosis well into the future. This review describes functional genomics approaches to apply the data generated by the Leptospira Genome Project towards deepening our knowledge of virulence factors of Leptospira using the emerging discipline of pathogenomics.
Figures



References
-
- Gouveia E.L., Metcalfe J., de Carvalho A.L., Aires T.S., Villasboas-Bisneto J.C., Queirroz A., Santos A.C., Salgado K., Reis M.G., Ko A.I. Leptospirosis-associated severe pulmonary hemorrhagic syndrome, Salvador, Brazil. Emerg. Infect. Dis. 2008;14:505–508. doi: 10.3201/eid1403.071064. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

