Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation
- PMID: 25437876
- PMCID: PMC4382238
- DOI: 10.1172/JCI76012
Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation
Abstract
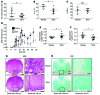
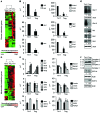
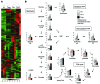
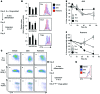
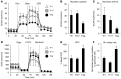
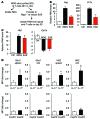
Activation of CD4+ T cells results in rapid proliferation and differentiation into effector and regulatory subsets. CD4+ effector T cell (Teff) (Th1 and Th17) and Treg subsets are metabolically distinct, yet the specific metabolic differences that modify T cell populations are uncertain. Here, we evaluated CD4+ T cell populations in murine models and determined that inflammatory Teffs maintain high expression of glycolytic genes and rely on high glycolytic rates, while Tregs are oxidative and require mitochondrial electron transport to proliferate, differentiate, and survive. Metabolic profiling revealed that pyruvate dehydrogenase (PDH) is a key bifurcation point between T cell glycolytic and oxidative metabolism. PDH function is inhibited by PDH kinases (PDHKs). PDHK1 was expressed in Th17 cells, but not Th1 cells, and at low levels in Tregs, and inhibition or knockdown of PDHK1 selectively suppressed Th17 cells and increased Tregs. This alteration in the CD4+ T cell populations was mediated in part through ROS, as N-acetyl cysteine (NAC) treatment restored Th17 cell generation. Moreover, inhibition of PDHK1 modulated immunity and protected animals against experimental autoimmune encephalomyelitis, decreasing Th17 cells and increasing Tregs. Together, these data show that CD4+ subsets utilize and require distinct metabolic programs that can be targeted to control specific T cell populations in autoimmune and inflammatory diseases.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

