PRIMUS: rapid reconstruction of pedigrees from genome-wide estimates of identity by descent
- PMID: 25439724
- PMCID: PMC4225580
- DOI: 10.1016/j.ajhg.2014.10.005
PRIMUS: rapid reconstruction of pedigrees from genome-wide estimates of identity by descent
Abstract
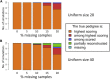
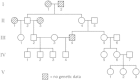
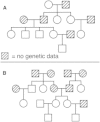
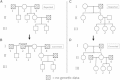
Understanding and correctly utilizing relatedness among samples is essential for genetic analysis; however, managing sample records and pedigrees can often be error prone and incomplete. Data sets ascertained by random sampling often harbor cryptic relatedness that can be leveraged in genetic analyses for maximizing power. We have developed a method that uses genome-wide estimates of pairwise identity by descent to identify families and quickly reconstruct and score all possible pedigrees that fit the genetic data by using up to third-degree relatives, and we have included it in the software package PRIMUS (Pedigree Reconstruction and Identification of the Maximally Unrelated Set). Here, we validate its performance on simulated, clinical, and HapMap pedigrees. Among these samples, we demonstrate that PRIMUS can verify reported pedigree structures and identify cryptic relationships. Finally, we show that PRIMUS reconstructed pedigrees, all of which were previously unknown, for 203 families from a cohort collected in Starr County, TX (1,890 samples).
Copyright © 2014 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
PRIMUS: improving pedigree reconstruction using mitochondrial and Y haplotypes.Bioinformatics. 2016 Feb 15;32(4):596-8. doi: 10.1093/bioinformatics/btv618. Epub 2015 Oct 29. Bioinformatics. 2016. PMID: 26515822 Free PMC article.
-
PADRE: Pedigree-Aware Distant-Relationship Estimation.Am J Hum Genet. 2016 Jul 7;99(1):154-62. doi: 10.1016/j.ajhg.2016.05.020. Epub 2016 Jun 30. Am J Hum Genet. 2016. PMID: 27374771 Free PMC article.
-
Profiling and Leveraging Relatedness in a Precision Medicine Cohort of 92,455 Exomes.Am J Hum Genet. 2018 May 3;102(5):874-889. doi: 10.1016/j.ajhg.2018.03.012. Am J Hum Genet. 2018. PMID: 29727688 Free PMC article.
-
Relatedness in the post-genomic era: is it still useful?Nat Rev Genet. 2015 Jan;16(1):33-44. doi: 10.1038/nrg3821. Epub 2014 Nov 18. Nat Rev Genet. 2015. PMID: 25404112 Review.
-
Wild pedigrees: the way forward.Proc Biol Sci. 2008 Mar 22;275(1635):613-21. doi: 10.1098/rspb.2007.1531. Proc Biol Sci. 2008. PMID: 18211868 Free PMC article. Review.
Cited by
-
Host genetic effects in pneumonia.Am J Hum Genet. 2021 Jan 7;108(1):194-201. doi: 10.1016/j.ajhg.2020.12.010. Epub 2020 Dec 13. Am J Hum Genet. 2021. PMID: 33357513 Free PMC article.
-
Large-scale multi-omics analyses in Hispanic/Latino populations identify genes for cardiometabolic traits.Nat Commun. 2025 Apr 11;16(1):3438. doi: 10.1038/s41467-025-58574-z. Nat Commun. 2025. PMID: 40210677 Free PMC article.
-
Comparison and assessment of family- and population-based genotype imputation methods in large pedigrees.Genome Res. 2019 Jan;29(1):125-134. doi: 10.1101/gr.236315.118. Epub 2018 Dec 4. Genome Res. 2019. PMID: 30514702 Free PMC article.
-
Bactrocera dorsalis in the Indian Ocean: A tale of two invasions.Evol Appl. 2022 Dec 1;16(1):48-61. doi: 10.1111/eva.13507. eCollection 2023 Jan. Evol Appl. 2022. PMID: 36699130 Free PMC article.
-
Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism.Cell. 2020 Feb 6;180(3):568-584.e23. doi: 10.1016/j.cell.2019.12.036. Epub 2020 Jan 23. Cell. 2020. PMID: 31981491 Free PMC article.
References
-
- Santorico S.A., Edwards K.L. Challenges of linkage analysis in the era of whole-genome sequencing. Genet. Epidemiol. 2014;38(Suppl 1):S92–S96. - PubMed
-
- Ott J., Kamatani Y., Lathrop M. Family-based designs for genome-wide association studies. Nat. Rev. Genet. 2011;12:465–474. - PubMed
-
- Below J.E., Earl D.L., Shively K.M., McMillin M.J., Smith J.D., Turner E.H., Stephan M.J., Al-Gazali L.I., Hertecant J.L., Chitayat D., University of Washington Center for Mendelian Genomics Whole-genome analysis reveals that mutations in inositol polyphosphate phosphatase-like 1 cause opsismodysplasia. Am. J. Hum. Genet. 2013;92:137–143. - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

