Dosage changes of a segment at 17p13.1 lead to intellectual disability and microcephaly as a result of complex genetic interaction of multiple genes
- PMID: 25439725
- PMCID: PMC4225592
- DOI: 10.1016/j.ajhg.2014.10.006
Dosage changes of a segment at 17p13.1 lead to intellectual disability and microcephaly as a result of complex genetic interaction of multiple genes
Abstract
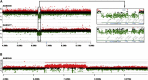
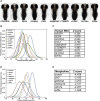
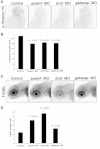
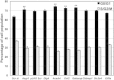
The 17p13.1 microdeletion syndrome is a recently described genomic disorder with a core clinical phenotype of intellectual disability, poor to absent speech, dysmorphic features, and a constellation of more variable clinical features, most prominently microcephaly. We identified five subjects with copy-number variants (CNVs) on 17p13.1 for whom we performed detailed clinical and molecular studies. Breakpoint mapping and retrospective analysis of published cases refined the smallest region of overlap (SRO) for microcephaly to a genomic interval containing nine genes. Dissection of this phenotype in zebrafish embryos revealed a complex genetic architecture: dosage perturbation of four genes (ASGR1, ACADVL, DVL2, and GABARAP) impeded neurodevelopment and decreased dosage of the same loci caused a reduced mitotic index in vitro. Moreover, epistatic analyses in vivo showed that dosage perturbations of discrete gene pairings induce microcephaly. Taken together, these studies support a model in which concomitant dosage perturbation of multiple genes within the CNV drive the microcephaly and possibly other neurodevelopmental phenotypes associated with rearrangements in the 17p13.1 SRO.
Copyright © 2014 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Lupski J.R., de Oca-Luna R.M., Slaugenhaupt S., Pentao L., Guzzetta V., Trask B.J., Saucedo-Cardenas O., Barker D.F., Killian J.M., Garcia C.A. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66:219–232. - PubMed
-
- Chance P.F., Alderson M.K., Leppig K.A., Lensch M.W., Matsunami N., Smith B., Swanson P.D., Odelberg S.J., Disteche C.M., Bird T.D. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72:143–151. - PubMed
-
- Potocki L., Bi W., Treadwell-Deering D., Carvalho C.M., Eifert A., Friedman E.M., Glaze D., Krull K., Lee J.A., Lewis R.A. Characterization of Potocki-Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. Am. J. Hum. Genet. 2007;80:633–649. - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

