DNA looping generated by DNA bending protein IHF and the two domains of lambda integrase
- PMID: 2544029
- PMCID: PMC1892171
- DOI: 10.1126/science.2544029
DNA looping generated by DNA bending protein IHF and the two domains of lambda integrase
Abstract
The multiprotein-DNA complexes that participate in bacteriophage lambda site-specific recombination were used to study the combined effect of protein-induced bending and protein-mediated looping of DNA. The protein integrase (Int) is a monomer with two autonomous DNA binding domains of different sequence specificity. Stimulation of Int binding and cleavage at the low affinity core-type DNA sites required interactions with the high affinity arm-type sites and depended on simultaneous binding of the sequence-specific DNA bending protein IHF (integration host factor). The bivalent DNA binding protein is positioned at high affinity sites and directed, by a DNA bending protein, to interactions with distant lower affinity sites. Assembly of this complex is independent of protein-protein interactions.
Figures


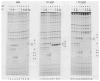
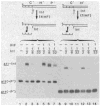

 ) and the end of a DNA fragment (
) and the end of a DNA fragment (
 ) are indicated. The pattern of enhanced binding for both the P′ arm and the core Int binding sites depends upon the presence or absence of adjacent DNA and other Int binding sites.
) are indicated. The pattern of enhanced binding for both the P′ arm and the core Int binding sites depends upon the presence or absence of adjacent DNA and other Int binding sites.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

