Long noncoding RNAs: an emerging link between gene regulation and nuclear organization
- PMID: 25441720
- PMCID: PMC4254690
- DOI: 10.1016/j.tcb.2014.08.009
Long noncoding RNAs: an emerging link between gene regulation and nuclear organization
Abstract
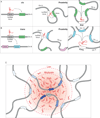
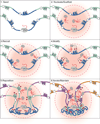
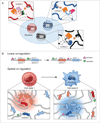
Mammalian genomes encode thousands of long noncoding RNAs (lncRNAs) that play important roles in diverse biological processes. As a class, lncRNAs are generally enriched in the nucleus and, specifically, within the chromatin-associated fraction. Consistent with their localization, many lncRNAs have been implicated in the regulation of gene expression and in shaping 3D nuclear organization. In this review, we discuss the evidence that many nuclear-retained lncRNAs can interact with various chromatin regulatory proteins and recruit them to specific sites on DNA to regulate gene expression. Furthermore, we discuss the role of specific lncRNAs in shaping nuclear organization and their emerging mechanisms. Based on these examples, we propose a model that explains how lncRNAs may shape aspects of nuclear organization to regulate gene expression.
Keywords: chromatin regulation; genome organization; long noncoding RNA (lncRNA); nuclear domains.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures


Similar articles
-
The Properties of Long Noncoding RNAs That Regulate Chromatin.Annu Rev Genomics Hum Genet. 2016 Aug 31;17:69-94. doi: 10.1146/annurev-genom-090314-024939. Epub 2016 Apr 21. Annu Rev Genomics Hum Genet. 2016. PMID: 27147088 Review.
-
Long non-coding RNAs and their functions in plants.Curr Opin Plant Biol. 2015 Oct;27:207-16. doi: 10.1016/j.pbi.2015.08.003. Epub 2015 Sep 3. Curr Opin Plant Biol. 2015. PMID: 26342908 Review.
-
Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression.Nat Rev Mol Cell Biol. 2016 Dec;17(12):756-770. doi: 10.1038/nrm.2016.126. Epub 2016 Oct 26. Nat Rev Mol Cell Biol. 2016. PMID: 27780979 Review.
-
Long noncoding RNAs as Organizers of Nuclear Architecture.Sci China Life Sci. 2016 Mar;59(3):236-44. doi: 10.1007/s11427-016-5012-y. Epub 2016 Jan 29. Sci China Life Sci. 2016. PMID: 26825945 Review.
-
Roles of long noncoding RNAs in chromosome domains.Wiley Interdiscip Rev RNA. 2017 Mar;8(2). doi: 10.1002/wrna.1384. Epub 2016 Aug 3. Wiley Interdiscip Rev RNA. 2017. PMID: 27489248 Review.
Cited by
-
The biological function and potential mechanism of long non-coding RNAs in cardiovascular disease.J Cell Mol Med. 2020 Nov;24(22):12900-12909. doi: 10.1111/jcmm.15968. Epub 2020 Oct 13. J Cell Mol Med. 2020. PMID: 33052009 Free PMC article. Review.
-
Angiogenesis-related non-coding RNAs and gastrointestinal cancer.Mol Ther Oncolytics. 2021 May 15;21:220-241. doi: 10.1016/j.omto.2021.04.002. eCollection 2021 Jun 25. Mol Ther Oncolytics. 2021. PMID: 34095461 Free PMC article. Review.
-
Long noncoding RNAs in cardiac development and ageing.Nat Rev Cardiol. 2015 Jul;12(7):415-25. doi: 10.1038/nrcardio.2015.55. Epub 2015 Apr 7. Nat Rev Cardiol. 2015. PMID: 25855606 Review.
-
Noncoding RNA MaIL1 is an integral component of the TLR4-TRIF pathway.Proc Natl Acad Sci U S A. 2020 Apr 21;117(16):9042-9053. doi: 10.1073/pnas.1920393117. Epub 2020 Apr 2. Proc Natl Acad Sci U S A. 2020. PMID: 32241891 Free PMC article.
-
Nucleolin facilitates nuclear retention of an ultraconserved region containing TRA2β4 and accelerates colon cancer cell growth.Oncotarget. 2018 Jun 1;9(42):26817-26833. doi: 10.18632/oncotarget.25510. eCollection 2018 Jun 1. Oncotarget. 2018. PMID: 29928487 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

