Diversity, expression and mRNA targeting abilities of Argonaute-targeting miRNAs among selected vascular plants
- PMID: 25443390
- PMCID: PMC4300679
- DOI: 10.1186/1471-2164-15-1049
Diversity, expression and mRNA targeting abilities of Argonaute-targeting miRNAs among selected vascular plants
Abstract
Background: Micro (mi)RNAs are important regulators of plant development. Across plant lineages, Dicer-like 1 (DCL1) proteins process long ds-like structures to produce micro (mi) RNA duplexes in a stepwise manner. These miRNAs are incorporated into Argonaute (AGO) proteins and influence expression of RNAs that have sequence complementarity with miRNAs. Expression levels of AGOs are greatly regulated by plants in order to minimize unwarranted perturbations using miRNAs to target mRNAs coding for AGOs. AGOs may also have high promoter specificity-sometimes expression of AGO can be limited to just a few cells in a plant. Viral pathogens utilize various means to counter antiviral roles of AGOs including hijacking the host encoded miRNAs to target AGOs. Two host encoded miRNAs namely miR168 and miR403 that target AGOs have been described in the model plant Arabidopsis and such a mechanism is thought to be well conserved across plants because AGO sequences are well conserved.
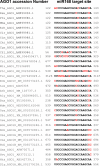
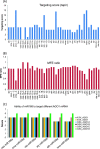
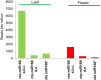
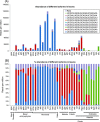
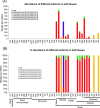
Results: We show that the interaction between AGO mRNAs and miRNAs is species-specific due to the diversity in sequences of two miRNAs that target AGOs, sequence diversity among corresponding target regions in AGO mRNAs and variable expression levels of these miRNAs among vascular plants. We used miRNA sequences from 68 plant species representing 31 plant families for this analysis. Sequences of miR168 and miR403 are not conserved among plant lineages, but surprisingly they differ drastically in their sequence diversity and expression levels even among closely related plants. Variation in miR168 expression among plants correlates well with secondary structures/length of loop sequences of their precursors.
Conclusions: Our data indicates a complex AGO targeting interaction among plant lineages due to miRNA sequence diversity and sequences of miRNA targeting regions among AGO mRNAs, thus leading to the assumption that the perturbations by viruses that use host miRNAs to target antiviral AGOs can only be species-specific. We also show that rapid evolution and likely loss of expression of miR168 isoforms in tobacco is related to the insertion of MITE-like transposons between miRNA and miRNA* sequences, a possible mechanism showing how miRNAs are lost in few plant lineages even though other close relatives have abundantly expressing miRNAs.
Figures


References
-
- Zhai J, Jeong D-H, De Paoli E, Park S, Rosen BD, Li Y, Gonzalez AJ, Yan Z, Kitto SL, Grusak MA, Jackson SA, Stacey G, Cook DR, Green PJ, Sherrier DJ, Meyers BC. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011;25:2540–2553. doi: 10.1101/gad.177527.111. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

