Recent advances of genomic testing in perinatal medicine
- PMID: 25444417
- PMCID: PMC4883661
- DOI: 10.1053/j.semperi.2014.10.009
Recent advances of genomic testing in perinatal medicine
Abstract
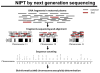
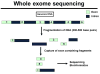
Rapid progress in genomic medicine in recent years has made it possible to diagnose subtle genetic abnormalities in a clinical setting on routine basis. This has allowed for detailed genotype-phenotype correlations and the identification of the genetic basis of many congenital anomalies. In addition to the availability of chromosomal microarray analysis, exome and whole-genome sequencing on pre- and postnatal samples of cell-free DNA has revolutionized the field of prenatal diagnosis. Incorporation of these technologies in perinatal pathology is bound to play a major role in coming years. In this communication, we briefly present the current experience with use of classical chromosome analysis, fluorescence in situ hybridization, and microarray testing, development of whole-genome analysis by next-generation sequencing technology, offer a detailed review of the history and current status of non-invasive prenatal testing using cell-free DNA, and discuss the advents of these new genomic technologies in perinatal medicine.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

