A moonlighting function of Mycobacterium smegmatis Ku in zinc homeostasis?
- PMID: 25450225
- PMCID: PMC4315663
- DOI: 10.1002/pro.2612
A moonlighting function of Mycobacterium smegmatis Ku in zinc homeostasis?
Abstract
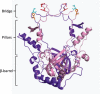
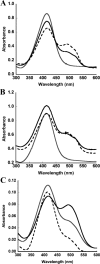
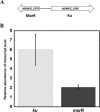
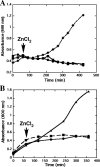
Ku protein participates in DNA double-strand break repair via the nonhomologous end-joining pathway. The three-dimensional structure of eukaryotic Ku reveals a central core consisting of a β-barrel domain and pillar and bridge regions that combine to form a ring-like structure that encircles DNA. Homologs of Ku are encoded by a subset of bacterial species, and they are predicted to conserve this core domain. In addition, the bridge region of Ku from some bacteria is predicted from homology modeling and sequence analyses to contain a conventional HxxC and CxxC (where x is any residue) zinc-binding motif. These potential zinc-binding sites have either deteriorated or been entirely lost in Ku from other organisms. Using an in vitro metal binding assay, we show that Mycobacterium smegmatis Ku binds two zinc ions. Zinc binding modestly stabilizes the Ku protein (by ∼3°C) and prevents cysteine oxidation, but it has little effect on DNA binding. In vivo, zinc induces significant upregulation of the gene encoding Ku (∼sixfold) as well as a divergently oriented gene encoding a predicted zinc-dependent MarR family transcription factor. Notably, overexpression of Ku confers zinc tolerance on Escherichia coli. We speculate that zinc-binding sites in Ku proteins from M. smegmatis and other mycobacterial species have been evolutionarily retained to provide protection against zinc toxicity without compromising the function of Ku in DNA double-strand break repair.
Keywords: DNA binding; Ku protein; MarR; PAR assay; thermal stability; zinc binding.
© 2014 The Protein Society.
Figures




References
-
- Doherty AJ, Jackson SP, Weller GR. Identification of bacterial homologues of the Ku DNA repair proteins. FEBS Lett. 2001;500:186–188. - PubMed
-
- Featherstone C, Jackson SP. Ku, a DNA repair protein with multiple cellular functions? Mutat Res. 1999;434:3–15. - PubMed
-
- Weller GR, Kysela B, Roy R, Tonkin LM, Scanlan E, Della M, Devine SK, Day JP, Wilkinson A, d'Adda di Fagagna F, Devine KM, Bowater RP, Jeggo PA, Jackson SP, Doherty AJ. Identification of a DNA nonhomologous end-joining complex in bacteria. Science. 2002;297:1686–1689. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

