The use of SMALPs as a novel membrane protein scaffold for structure study by negative stain electron microscopy
- PMID: 25450810
- PMCID: PMC4331651
- DOI: 10.1016/j.bbamem.2014.10.018
The use of SMALPs as a novel membrane protein scaffold for structure study by negative stain electron microscopy
Abstract
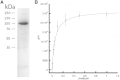
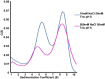
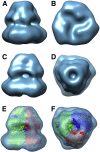
Despite the great progress recently made in resolving their structures, investigation of the structural biology of membrane proteins still presents major challenges. Even with new technical advances such as lipidic cubic phase crystallisation, obtaining well-ordered crystals remains a significant hurdle in membrane protein X-ray crystallographic studies. As an alternative, electron microscopy has been shown to be capable of resolving >3.5Å resolution detail in membrane proteins of modest (~300 kDa) size, without the need for crystals. However, the conventional use of detergents for either approach presents several issues, including the possible effects on structure of removing the proteins from their natural membrane environment. As an alternative, it has recently been demonstrated that membrane proteins can be effectively isolated, in the absence of detergents, using a styrene maleic acid co-polymer (SMA). This approach yields SMA lipid particles (SMALPs) in which the membrane proteins are surrounded by a small disk of lipid bilayer encircled by polymer. Here we use the Escherichia coli secondary transporter AcrB as a model membrane protein to demonstrate how a SMALP scaffold can be used to visualise membrane proteins, embedded in a near-native lipid environment, by negative stain electron microscopy, yielding structures at a modest resolution in a short (days) timeframe. Moreover, we show that AcrB within a SMALP scaffold is significantly more active than the equivalent DDM stabilised form. The advantages of SMALP scaffolds within electron microscopy are discussed and we conclude that they may prove to be an important tool in studying membrane protein structure and function.
Keywords: AcrB; Electron microscopy; Membrane protein; SMALP.
Copyright © 2014. Published by Elsevier B.V.
Figures


References
-
- Terstappen G.C., Roncarati R., Dunlop J., Peri R. Screening technologies for ion channel drug discovery. Future Med. Chem. 2010;2:715–730. - PubMed
-
- Postis V.L., Deacon S.E., Roach P.C., Wright G.S., Xia X., Ingram J.C., Hadden J.M., Henderson P.J., Phillips S.E., McPherson M.J., Baldwin S.A. A high-throughput assay of membrane protein stability. Mol. Membr. Biol. 2008;25:617–624. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

