An updated view on the structure and function of PYRIN domains
- PMID: 25451010
- PMCID: PMC4297229
- DOI: 10.1007/s10495-014-1065-1
An updated view on the structure and function of PYRIN domains
Abstract
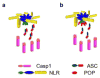
The PYRIN domain (PYD) is a protein-protein interaction domain, which belongs to the death domain fold (DDF) superfamily. It is best known for its signaling function in innate immune responses and particularly in the assembly of inflammasomes, which are large protein complexes that allow the induced proximity-mediated activation of caspase-1 and subsequently the release of pro-inflammatory cytokines. The molecular mechanism of inflammasome assembly was only recently elucidated and specifically requires PYD oligomerization. Here we discuss the recent advances in our understanding of PYD signaling and its regulation by PYD-only proteins.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






References
-
- Ashkenazi A, Salvesen G. Regulated Cell Death: Signaling and Mechanisms. Annu Rev Cell Dev Biol. 2014;30:337–356. - PubMed
-
- Boatright KM, Salvesen GS. Mechanisms of caspase activation. Current Opinion in Cell Biology. 2003;15:725–731. - PubMed
-
- Chang DW, Ditsworth D, Liu H, Srinivasula SM, Alnemri ES, Yang X. Oligomerization is a general mechanism for the activation of apoptosis initiator and inflammatory procaspases. J Biol Chem. 2003;278:16466–16469. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

