The high-risk HPV16 E7 oncoprotein mediates interaction between the transcriptional coactivator CBP and the retinoblastoma protein pRb
- PMID: 25451029
- PMCID: PMC4258470
- DOI: 10.1016/j.jmb.2014.10.021
The high-risk HPV16 E7 oncoprotein mediates interaction between the transcriptional coactivator CBP and the retinoblastoma protein pRb
Abstract
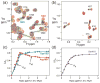
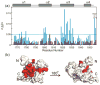
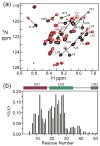
The oncoprotein E7 from human papillomavirus (HPV) strains that confer high cancer risk mediates cell transformation by deregulating host cellular processes and activating viral gene expression through recruitment of cellular proteins such as the retinoblastoma protein (pRb) and the cyclic-AMP response element binding binding protein (CBP) and its paralog p300. Here we show that the intrinsically disordered N-terminal region of E7 from high-risk HPV16 binds the TAZ2 domain of CBP with greater affinity than E7 from low-risk HPV6b. HPV E7 and the tumor suppressor p53 compete for binding to TAZ2. The TAZ2 binding site in E7 overlaps the LxCxE motif that is crucial for interaction with pRb. While TAZ2 and pRb compete for binding to a monomeric E7 polypeptide, the full-length E7 dimer mediates an interaction between TAZ2 and pRb by promoting formation of a ternary complex. Cell-based assays show that expression of full-length HPV16 E7 promotes increased pRb acetylation and that this response depends both on the presence of CBP/p300 and on the ability of E7 to form a dimer. These observations suggest a model for the oncogenic effect of high-risk HPV16 E7. The disordered region of one E7 molecule in the homodimer interacts with the pocket domain of pRb, while the same region of the other E7 molecule binds the TAZ2 domain of CBP/p300. Through its ability to dimerize, E7 recruits CBP/p300 and pRb into a ternary complex, bringing the histone acetyltransferase domain of CBP/p300 into proximity to pRb and promoting acetylation, leading to disruption of cell cycle control.
Keywords: CBP/p300; NMR; acetyltransferase; human papillomavirus; intrinsic protein disorder.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures

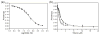





References
-
- de Villiers E-M, Fauquet C, Broker TR, Bernard H-U, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. - PubMed
-
- zur Hausen H. Papillomavirus infections--a major cause of human cancers. Biochim Biophys Acta. 1996;1288:F55–78. - PubMed
-
- zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nature Rev Cancer. 2002;2:342–50. - PubMed
-
- Berk AJ. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene. 2005;24:7673–85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

