Regulation of iNOS function and cellular redox state by macrophage Gch1 reveals specific requirements for tetrahydrobiopterin in NRF2 activation
- PMID: 25451639
- PMCID: PMC4344222
- DOI: 10.1016/j.freeradbiomed.2014.10.575
Regulation of iNOS function and cellular redox state by macrophage Gch1 reveals specific requirements for tetrahydrobiopterin in NRF2 activation
Abstract
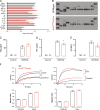
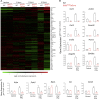
Inducible nitric oxide synthase (iNOS) is a key enzyme in the macrophage inflammatory response, which is the source of nitric oxide (NO) that is potently induced in response to proinflammatory stimuli. However, the specific role of NO production, as distinct from iNOS induction, in macrophage inflammatory responses remains unproven. We have generated a novel mouse model with conditional deletion of Gch1, encoding GTP cyclohydrolase 1 (GTPCH), an essential enzyme in the biosynthesis of tetrahydrobiopterin (BH4) that is a required cofactor for iNOS NO production. Mice with a floxed Gch1 allele (Gch1(fl/fl)) were crossed with Tie2cre transgenic mice, causing Gch1 deletion in leukocytes (Gch1(fl/fl)Tie2cre). Macrophages from Gch1(fl/fl)Tie2cre mice lacked GTPCH protein and de novo biopterin biosynthesis. When activated with LPS and IFNγ, macrophages from Gch1(fl/fl)Tie2cre mice induced iNOS protein in a manner indistinguishable from wild-type controls, but produced no detectable NO, as judged by L-citrulline production, EPR spin trapping of NO, and by nitrite accumulation. Incubation of Gch1(fl/fl)Tie2cre macrophages with dihydroethidium revealed significantly increased production of superoxide in the presence of iNOS expression, and an iNOS-independent, BH4-dependent increase in other ROS species. Normal BH4 levels, nitric oxide production, and cellular redox state were restored by sepiapterin, a precursor of BH4 production by the salvage pathway, demonstrating that the effects of BH4 deficiency were reversible. Gch1(fl/fl)Tie2cre macrophages showed only minor alterations in cytokine production and normal cell migration, and minimal changes in basal gene expression. However, gene expression analysis after iNOS induction identified 78 genes that were altered between wild-type and Gch1(fl/fl)Tie2cre macrophages. Pathway analysis identified decreased NRF2 activation, with reduced induction of archetypal NRF2 genes (gclm, prdx1, gsta3, nqo1, and catalase) in BH4-deficient Gch1(fl/fl)Tie2cre macrophages. These findings identify BH4-dependent iNOS regulation and NO generation as specific requirements for NRF2-dependent responses in macrophage inflammatory activation.
Keywords: As3MT; BH4; GCH1; GTPCH; Macrophage; NOS2; Nitric oxide; Tetrahydrobiopterin; iNOS.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





Similar articles
-
A key role for tetrahydrobiopterin-dependent endothelial NOS regulation in resistance arteries: studies in endothelial cell tetrahydrobiopterin-deficient mice.Br J Pharmacol. 2017 Apr;174(8):657-671. doi: 10.1111/bph.13728. Epub 2017 Mar 13. Br J Pharmacol. 2017. PMID: 28128438 Free PMC article.
-
Cell-autonomous role of endothelial GTP cyclohydrolase 1 and tetrahydrobiopterin in blood pressure regulation.Hypertension. 2014 Sep;64(3):530-40. doi: 10.1161/HYPERTENSIONAHA.114.03089. Epub 2014 Apr 28. Hypertension. 2014. PMID: 24777984 Free PMC article.
-
Regulation of mycobacterial infection by macrophage Gch1 and tetrahydrobiopterin.Nat Commun. 2018 Dec 20;9(1):5409. doi: 10.1038/s41467-018-07714-9. Nat Commun. 2018. PMID: 30573728 Free PMC article.
-
Catecholamines and Parkinson's disease: tyrosine hydroxylase (TH) over tetrahydrobiopterin (BH4) and GTP cyclohydrolase I (GCH1) to cytokines, neuromelanin, and gene therapy: a historical overview.J Neural Transm (Vienna). 2024 Jun;131(6):617-630. doi: 10.1007/s00702-023-02673-y. Epub 2023 Aug 28. J Neural Transm (Vienna). 2024. PMID: 37638996 Review.
-
Tetrahydrobioterin (BH4) Pathway: From Metabolism to Neuropsychiatry.Curr Neuropharmacol. 2021;19(5):591-609. doi: 10.2174/1570159X18666200729103529. Curr Neuropharmacol. 2021. PMID: 32744952 Free PMC article. Review.
Cited by
-
Repair and regeneration: ferroptosis in the process of remodeling and fibrosis in impaired organs.Cell Death Discov. 2024 Oct 2;10(1):424. doi: 10.1038/s41420-024-02181-2. Cell Death Discov. 2024. PMID: 39358326 Free PMC article. Review.
-
Sophoridine Inhibits the Tumour Growth of Non-Small Lung Cancer by Inducing Macrophages M1 Polarisation via MAPK-Mediated Inflammatory Pathway.Front Oncol. 2021 Feb 24;11:634851. doi: 10.3389/fonc.2021.634851. eCollection 2021. Front Oncol. 2021. PMID: 33718223 Free PMC article.
-
BH4 supplementation reduces retinal cell death in ischaemic retinopathy.Sci Rep. 2023 Dec 2;13(1):21292. doi: 10.1038/s41598-023-48167-5. Sci Rep. 2023. PMID: 38042898 Free PMC article.
-
Phosphatidylinositide 3-Kinase Contributes to the Anti-Inflammatory Effect of Abutilon crispum L. Medik Methanol Extract.Evid Based Complement Alternat Med. 2018 Nov 26;2018:1935902. doi: 10.1155/2018/1935902. eCollection 2018. Evid Based Complement Alternat Med. 2018. PMID: 30598682 Free PMC article.
-
Global analysis of erythroid cells redox status reveals the involvement of Prdx1 and Prdx2 in the severity of beta thalassemia.PLoS One. 2018 Dec 6;13(12):e0208316. doi: 10.1371/journal.pone.0208316. eCollection 2018. PLoS One. 2018. PMID: 30521599 Free PMC article.
References
-
- MacMicking J.D., Nathan C., Hom G., Chartrain N., Fletcher D.S., Trumbauer M., Stevens K., Xie Q., Sokol K., Hutchinson N., Chen H., Mudgett J.S. Altered responses to bacterial infection and endotoxic shock in mice lacking inducible nitric oxide synthase. Cell. 1995;81:641–650. - PubMed
-
- Ishii T., Itoh K., Ruiz E., Leake D.S., Unoki H., Yamamoto M., Mann G.E. Role of Nrf2 in the regulation of CD36 and stress protein expression in murine macrophages: activation by oxidatively modified LDL and 4-hydroxynonenal. Circ. Res. 2004;94:609–616. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

