Mechanistic characterization of the thioredoxin system in the removal of hydrogen peroxide
- PMID: 25451645
- PMCID: PMC4280359
- DOI: 10.1016/j.freeradbiomed.2014.10.508
Mechanistic characterization of the thioredoxin system in the removal of hydrogen peroxide
Abstract
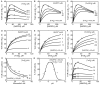
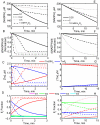
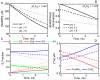
The thioredoxin system, which consists of a family of proteins, including thioredoxin (Trx), peroxiredoxin (Prx), and thioredoxin reductase (TrxR), plays a critical role in the defense against oxidative stress by removing harmful hydrogen peroxide (H2O2). Specifically, Trx donates electrons to Prx to remove H2O2 and then TrxR maintains the reduced Trx concentration with NADPH as the cofactor. Despite a great deal of kinetic information gathered on the removal of H2O2 by the Trx system from various sources/species, a mechanistic understanding of the associated enzymes is still not available. We address this issue by developing a thermodynamically consistent mathematical model of the Trx system which entails mechanistic details and provides quantitative insights into the kinetics of the TrxR and Prx enzymes. Consistent with experimental studies, the model analyses of the available data show that both enzymes operate by a ping-pong mechanism. The proposed mechanism for TrxR, which incorporates substrate inhibition by NADPH and intermediate protonation states, well describes the available data and accurately predicts the bell-shaped behavior of the effect of pH on the TrxR activity. Most importantly, the model also predicts the inhibitory effects of the reaction products (NADP(+) and Trx(SH)2) on the TrxR activity for which suitable experimental data are not available. The model analyses of the available data on the kinetics of Prx from mammalian sources reveal that Prx operates at very low H2O2 concentrations compared to their human parasite counterparts. Furthermore, the model is able to predict the dynamic overoxidation of Prx at high H2O2 concentrations, consistent with the available data. The integrated Prx-TrxR model simulations well describe the NADPH and H2O2 degradation dynamics and also show that the coupling of TrxR- and Prx-dependent reduction of H2O2 allowed ultrasensitive changes in the Trx concentration in response to changes in the TrxR concentration at high Prx concentrations. Thus, the model of this sort is very useful for integration into computational H2O2 degradation models to identify its role in physiological and pathophysiological functions.
Keywords: Enzyme kinetics; Hydrogen peroxide; Mathematical model; ROS scavenging; Redox biology; Thioredoxin system.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures





References
-
- Kamata H, Hirata H. Redox regulation of cellular signalling. Cellular Signalling. 1999;11:1–14. - PubMed
-
- Rhee SG, Yang KS, Kang SW, Woo HA, Chang TS. Controlled elimination of intracellular H2O2: Regulation of peroxiredoxin, catalase, and glutathione peroxidase via post-translational modification. Antioxidants and Redox Signaling. 2005;7:619–626. - PubMed
-
- Veal EA, Day AM, Morgan BA. Hydrogen peroxide sensing and signaling. Molecular Cell. 2007;26:1–14. - PubMed
-
- Andreyev AY, Kushnareva YE, Starkov AA. Mitochondrial metabolism of reactive oxygen species. Biochemistry (Moscow) 2005;70:200–214. - PubMed
-
- Brand MD, Affourtit C, Esteves TC, Green K, Lambert AJ, Miwa S, Pakay JL, Parker N. Mitochondrial superoxide: Production, biological effects, and activation of uncoupling proteins. Free Radical Biology and Medicine. 2004;37:755–767. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

