Inflammatory cytokines epigenetically regulate rheumatoid arthritis fibroblast-like synoviocyte activation by suppressing HDAC5 expression
- PMID: 25452308
- PMCID: PMC5336378
- DOI: 10.1136/annrheumdis-2014-205635
Inflammatory cytokines epigenetically regulate rheumatoid arthritis fibroblast-like synoviocyte activation by suppressing HDAC5 expression
Abstract
Objectives: Epigenetic modifications play an important role in the regulation of gene transcription and cellular function. Here, we examined if pro-inflammatory factors present in the inflamed joint of patients with rheumatoid arthritis (RA) could regulate histone deacetylase (HDAC) expression and function in fibroblast-like synoviocytes (FLS).
Methods: Protein acetylation in synovial tissue was assessed by immunohistochemistry. The mRNA levels of HDAC family members and inflammatory mediators in the synovial tissue and the changes in HDAC expression in RA FLS were measured by quantitative (q) PCR. FLS were either transfected with HDAC5 siRNA or transduced with adenoviral vector encoding wild-type HDAC5 and the effects of HDAC5 manipulation were examined by qPCR arrays, ELISA and ELISA-based assays.
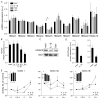
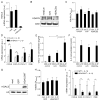
Results: Synovial class I HDAC expression was associated with local expression of tumour necrosis factor (TNF) and matrix metalloproteinase-1, while class IIa HDAC5 expression was inversely associated with parameters of disease activity (erythrocyte sedimentation rate, C-reactive protein, Disease Activity Score in 28 Joints). Interleukin (IL)-1β or TNF stimulation selectively suppressed HDAC5 expression in RA FLS, which was sufficient and required for optimal IFNB, CXCL9, CXCL10 and CXCL11 induction by IL-1β, associated with increased nuclear accumulation of the transcription factor, interferon regulatory factor 1(IRF1).
Conclusions: Inflammatory cytokines suppress RA FLS HDAC5 expression, promoting nuclear localisation of IRF1 and transcription of a subset of type I interferon response genes. Our results identify HDAC5 as a novel inflammatory mediator in RA, and suggest that strategies rescuing HDAC5 expression in vivo, or the development of HDAC inhibitors not affecting HDAC5 activity, may have therapeutic applications in RA treatment.
Keywords: Chemokines; Fibroblasts; Inflammation; Rheumatoid Arthritis.
Published by the BMJ Publishing Group Limited. For permission to use (where not already granted under a licence) please go to http://www.bmj.com/company/products-services/rights-and-licensing/
Conflict of interest statement
Figures



References
-
- Costenbader KH, Gay S, Alarcon-Riquelme ME, et al. Genes, epigenetic regulation and environmental factors: which is the most relevant in developing autoimmune diseases? Autoimmun Rev. 2012;11:604–9. - PubMed
-
- Grabiec AM, Reedquist KA. The ascent of acetylation in the epigenetics of rheumatoid arthritis. Nat Rev Rheumatol. 2013;9:311–18. - PubMed
-
- Ospelt C, Reedquist KA, Gay S, et al. Inflammatory memories: is epigenetics the missing link to persistent stromal cell activation in rheumatoid arthritis? Autoimmun Rev. 2011;10:519–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

