Epigenetic regulation in plants
- PMID: 25452385
- PMCID: PMC4292151
- DOI: 10.1101/cshperspect.a019315
Epigenetic regulation in plants
Abstract
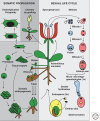
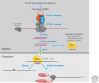
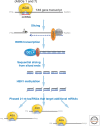
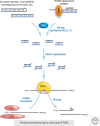
The study of epigenetics in plants has a long and rich history, from initial descriptions of non-Mendelian gene behaviors to seminal discoveries of chromatin-modifying proteins and RNAs that mediate gene silencing in most eukaryotes, including humans. Genetic screens in the model plant Arabidopsis have been particularly rewarding, identifying more than 130 epigenetic regulators thus far. The diversity of epigenetic pathways in plants is remarkable, presumably contributing to the phenotypic plasticity of plant postembryonic development and the ability to survive and reproduce in unpredictable environments.
Copyright © 2014 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures


References
WWW RESOURCES
-
- http://www.arabidopsis.leeds.ac.uk/act/coexpanalyser.php Arabidopsis coexpression mining.
-
- http://asrp.cgrb.oregonstate.edu Arabidopsis small RNA.
-
- http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi Gene expression in Arabidopsis.
-
- http://www.chromdb.org Chromatin genes.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
