SOXC proteins amplify canonical WNT signaling to secure nonchondrocytic fates in skeletogenesis
- PMID: 25452386
- PMCID: PMC4259807
- DOI: 10.1083/jcb.201405098
SOXC proteins amplify canonical WNT signaling to secure nonchondrocytic fates in skeletogenesis
Abstract
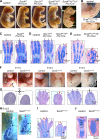
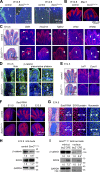
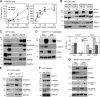
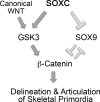
Canonical WNT signaling stabilizes β-catenin to determine cell fate in many processes from development onwards. One of its main roles in skeletogenesis is to antagonize the chondrogenic transcription factor SOX9. We here identify the SOXC proteins as potent amplifiers of this pathway. The SOXC genes, i.e., Sox4, Sox11, and Sox12, are coexpressed in skeletogenic mesenchyme, including presumptive joints and perichondrium, but not in cartilage. Their inactivation in mouse embryo limb bud caused massive cartilage fusions, as joint and perichondrium cells underwent chondrogenesis. SOXC proteins govern these cells cell autonomously. They replace SOX9 in the adenomatous polyposis coli-Axin destruction complex and therein inhibit phosphorylation of β-catenin by GSK3. This inhibition, a crucial, limiting step in canonical WNT signaling, thus becomes a constitutive event. The resulting SOXC/canonical WNT-mediated synergistic stabilization of β-catenin contributes to efficient repression of Sox9 in presumptive joint and perichondrium cells and thereby ensures proper delineation and articulation of skeletal primordia. This synergy may determine cell fate in many processes besides skeletogenesis.
© 2014 Bhattaram et al.
Figures




References
-
- Akiyama H., Chaboissier M.C., Martin J.F., Schedl A., and de Crombrugghe B.. 2002. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 16:2813–2828 10.1101/gad.1017802 - DOI - PMC - PubMed
-
- Albrecht U., Eichele G., Helms J.A., and Lu H.C.. 1997. Visualization of gene expression patterns by in situ hybridization. Molecular and Cellular Methods in Developmental Toxicology. Daston G.P., editor CRC Press, Inc., Boca Raton, FL: 23–48.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

