The molecular bases of δ/αβ T cell-mediated antigen recognition
- PMID: 25452463
- PMCID: PMC4267242
- DOI: 10.1084/jem.20141764
The molecular bases of δ/αβ T cell-mediated antigen recognition
Abstract
αβ and γδ T cells are disparate T cell lineages that can respond to distinct antigens (Ags) via the use of the αβ and γδ T cell Ag receptors (TCRs), respectively. Here we characterize a population of human T cells, which we term δ/αβ T cells, expressing TCRs comprised of a TCR-δ variable gene (Vδ1) fused to joining α and constant α domains, paired with an array of TCR-β chains. We demonstrate that these cells, which represent ∼50% of all Vδ1(+) human T cells, can recognize peptide- and lipid-based Ags presented by human leukocyte antigen (HLA) and CD1d, respectively. Similar to type I natural killer T (NKT) cells, CD1d-lipid Ag-reactive δ/αβ T cells recognized α-galactosylceramide (α-GalCer); however, their fine specificity for other lipid Ags presented by CD1d, such as α-glucosylceramide, was distinct from type I NKT cells. Thus, δ/αβTCRs contribute new patterns of Ag specificity to the human immune system. Furthermore, we provide the molecular bases of how δ/αβTCRs bind to their targets, with the Vδ1-encoded region providing a major contribution to δ/αβTCR binding. Our findings highlight how components from αβ and γδTCR gene loci can recombine to confer Ag specificity, thus expanding our understanding of T cell biology and TCR diversity.
© 2014 Pellicci et al.
Figures



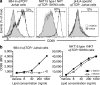

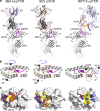
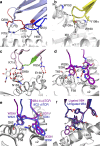
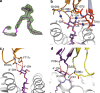

References
-
- Amir A.L., van der Steen D.M., Hagedoorn R.S., Kester M.G., van Bergen C.A., Drijfhout J.W., de Ru A.H., Falkenburg J.H., van Veelen P.A., and Heemskerk M.H.. 2011. Allo-HLA-reactive T cells inducing graft-versus-host disease are single peptide specific. Blood. 118:6733–6742 10.1182/blood-2011-05-354787 - DOI - PubMed
-
- Borg N.A., Ely L.K., Beddoe T., Macdonald W.A., Reid H.H., Clements C.S., Purcell A.W., Kjer-Nielsen L., Miles J.J., Burrows S.R., et al. . 2005. The CDR3 regions of an immunodominant T cell receptor dictate the ‘energetic landscape’ of peptide-MHC recognition. Nat. Immunol. 6:171–180 10.1038/ni1155 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

