Vibrio effector protein VopQ inhibits fusion of V-ATPase-containing membranes
- PMID: 25453092
- PMCID: PMC4291640
- DOI: 10.1073/pnas.1413764111
Vibrio effector protein VopQ inhibits fusion of V-ATPase-containing membranes
Abstract
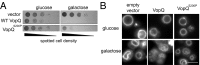
Vesicle fusion governs many important biological processes, and imbalances in the regulation of membrane fusion can lead to a variety of diseases such as diabetes and neurological disorders. Here we show that the Vibrio parahaemolyticus effector protein VopQ is a potent inhibitor of membrane fusion based on an in vitro yeast vacuole fusion model. Previously, we demonstrated that VopQ binds to the V(o) domain of the conserved V-type H(+)-ATPase (V-ATPase) found on acidic compartments such as the yeast vacuole. VopQ forms a nonspecific, voltage-gated membrane channel of 18 Å resulting in neutralization of these compartments. We now present data showing that VopQ inhibits yeast vacuole fusion. Furthermore, we identified a unique mutation in VopQ that delineates its two functions, deacidification and inhibition of membrane fusion. The use of VopQ as a membrane fusion inhibitor in this manner now provides convincing evidence that vacuole fusion occurs independently of luminal acidification in vitro.
Keywords: SNARE; Vibrio parahaemolyticus; vesicle fusion; vp1680; yeast vacuole.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Comment in
-
What are the roles of V-ATPases in membrane fusion?Proc Natl Acad Sci U S A. 2015 Jan 6;112(1):8-9. doi: 10.1073/pnas.1422280112. Epub 2014 Dec 24. Proc Natl Acad Sci U S A. 2015. PMID: 25540413 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

