EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma
- PMID: 25453903
- PMCID: PMC4492343
- DOI: 10.1016/j.ccell.2014.10.004
EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma
Abstract
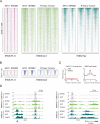
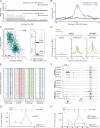
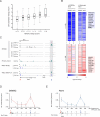
The aberrant transcription factor EWS-FLI1 drives Ewing sarcoma, but its molecular function is not completely understood. We find that EWS-FLI1 reprograms gene regulatory circuits in Ewing sarcoma by directly inducing or repressing enhancers. At GGAA repeat elements, which lack evolutionary conservation and regulatory potential in other cell types, EWS-FLI1 multimers induce chromatin opening and create de novo enhancers that physically interact with target promoters. Conversely, EWS-FLI1 inactivates conserved enhancers containing canonical ETS motifs by displacing wild-type ETS transcription factors. These divergent chromatin-remodeling patterns repress tumor suppressors and mesenchymal lineage regulators while activating oncogenes and potential therapeutic targets, such as the kinase VRK1. Our findings demonstrate how EWS-FLI1 establishes an oncogenic regulatory program governing both tumor survival and differentiation.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures




Comment in
-
ChIP-ping away at EWS/ETS transcription networks.Cancer Cell. 2014 Nov 10;26(5):595-6. doi: 10.1016/j.ccell.2014.10.016. Epub 2014 Nov 10. Cancer Cell. 2014. PMID: 25517742
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
