Influenza A virus transmission bottlenecks are defined by infection route and recipient host
- PMID: 25456074
- PMCID: PMC4272616
- DOI: 10.1016/j.chom.2014.09.020
Influenza A virus transmission bottlenecks are defined by infection route and recipient host
Abstract
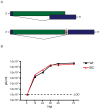
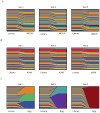
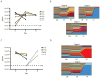
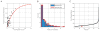
Despite its global relevance, our understanding of how influenza A virus transmission impacts the overall population dynamics of this RNA virus remains incomplete. To define this dynamic, we inserted neutral barcodes into the influenza A virus genome to generate a population of viruses that can be individually tracked during transmission events. We find that physiological bottlenecks differ dramatically based on the infection route and level of adaptation required for efficient replication. Strong genetic pressures are responsible for bottlenecks during adaptation across different host species, whereas transmission between susceptible hosts results in bottlenecks that are not genetically driven and occur at the level of the recipient. Additionally, the infection route significantly influences the bottleneck stringency, with aerosol transmission imposing greater selection than direct contact. These transmission constraints have implications in understanding the global migration of virus populations and provide a clearer perspective on the emergence of pandemic strains.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Barcoding influenza virus to decode transmission.Cell Host Microbe. 2014 Nov 12;16(5):559-61. doi: 10.1016/j.chom.2014.10.016. Epub 2014 Nov 12. Cell Host Microbe. 2014. PMID: 25525788
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

